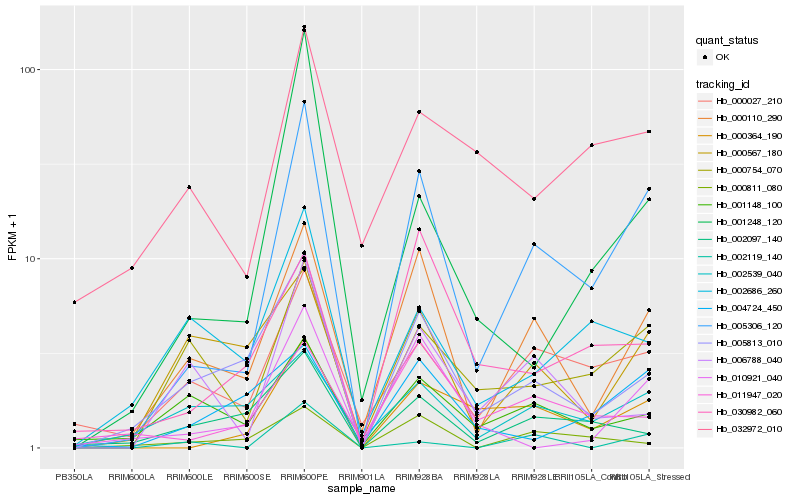
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005306_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000110_290 |
0.1953605121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000364_190 |
0.2006836624 |
- |
- |
- |
| 4 |
Hb_000754_070 |
0.2357505569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000027_210 |
0.2391530946 |
- |
- |
Os01g0689300, partial [Oryza sativa Japonica Group] |
| 6 |
Hb_005813_010 |
0.2477265407 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001248_120 |
0.2629937151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010921_040 |
0.2677293964 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 9 |
Hb_006788_040 |
0.2757221204 |
- |
- |
calmodulin, putative [Ricinus communis] |
| 10 |
Hb_032972_010 |
0.2825227024 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 11 |
Hb_011947_020 |
0.2836924567 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 12 |
Hb_001148_100 |
0.2895789251 |
- |
- |
- |
| 13 |
Hb_002686_260 |
0.2897713739 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 14 |
Hb_004724_450 |
0.2899666622 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 15 |
Hb_030982_060 |
0.2905674798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000811_080 |
0.2916568225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002119_140 |
0.2922321093 |
- |
- |
PREDICTED: uncharacterized protein LOC105634172 [Jatropha curcas] |
| 18 |
Hb_002539_040 |
0.2924176419 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 19 |
Hb_000567_180 |
0.2945740778 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 20 |
Hb_002097_140 |
0.2956781683 |
- |
- |
CHY zinc finger family protein, expressed [Oryza sativa Japonica Group] |