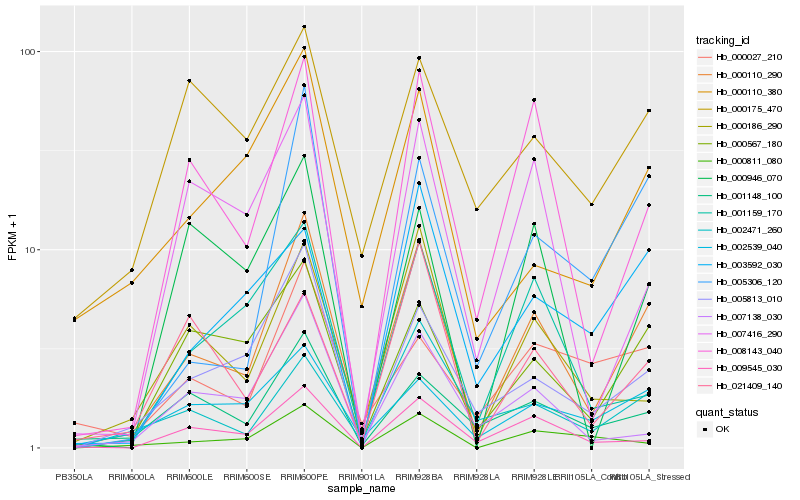
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_290 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008143_040 |
0.1751958233 |
- |
- |
Alpha-expansin 4 precursor, putative [Ricinus communis] |
| 3 |
Hb_000567_180 |
0.1812993851 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 4 |
Hb_005813_010 |
0.1877408369 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_005306_120 |
0.1953605121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_009545_030 |
0.2021593354 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000186_290 |
0.2047685163 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001148_100 |
0.2100801896 |
- |
- |
- |
| 9 |
Hb_002539_040 |
0.2112453297 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 10 |
Hb_000110_380 |
0.2120795085 |
- |
- |
PREDICTED: uncharacterized protein LOC105642019 [Jatropha curcas] |
| 11 |
Hb_007138_030 |
0.2129473315 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 12 |
Hb_007416_290 |
0.2149050209 |
- |
- |
PREDICTED: plasma membrane ATPase 4-like [Pyrus x bretschneideri] |
| 13 |
Hb_002471_260 |
0.2190177413 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 14 |
Hb_001159_170 |
0.2210161557 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 15 |
Hb_000811_080 |
0.2258321857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003592_030 |
0.2281420956 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 17 |
Hb_021409_140 |
0.2287506937 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 18 |
Hb_000175_470 |
0.2310880205 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 19 |
Hb_000946_070 |
0.2312592082 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 20 |
Hb_000027_210 |
0.2314377815 |
- |
- |
Os01g0689300, partial [Oryza sativa Japonica Group] |