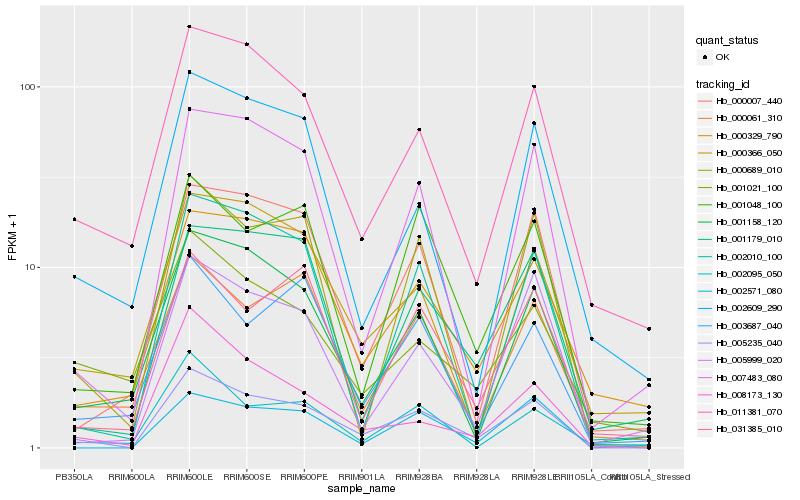
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005235_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 2 |
Hb_011381_070 |
0.157619485 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 3 |
Hb_001158_120 |
0.162496448 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007483_080 |
0.1639191991 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 5 |
Hb_001048_100 |
0.1706031577 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 6 |
Hb_002571_080 |
0.1719563326 |
- |
- |
secondary cell wall-related glycosyltransferase family 14 [Populus tremula x Populus tremuloides] |
| 7 |
Hb_000007_440 |
0.1720844539 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 8 |
Hb_000689_010 |
0.1725395539 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000329_790 |
0.1734138181 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 10 |
Hb_001179_010 |
0.1757822449 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 11 |
Hb_002010_100 |
0.1758475864 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 12 |
Hb_000061_310 |
0.1769654726 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005999_020 |
0.1778296276 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 14 |
Hb_031385_010 |
0.178167582 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 15 |
Hb_000366_050 |
0.1805856552 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_008173_130 |
0.1840473115 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003687_040 |
0.1850709171 |
- |
- |
PREDICTED: uncharacterized protein LOC105632153 [Jatropha curcas] |
| 18 |
Hb_001021_100 |
0.1860552416 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 19 |
Hb_002609_290 |
0.1895502941 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 20 |
Hb_002095_050 |
0.1922963005 |
- |
- |
PREDICTED: uncharacterized protein LOC104447479 [Eucalyptus grandis] |