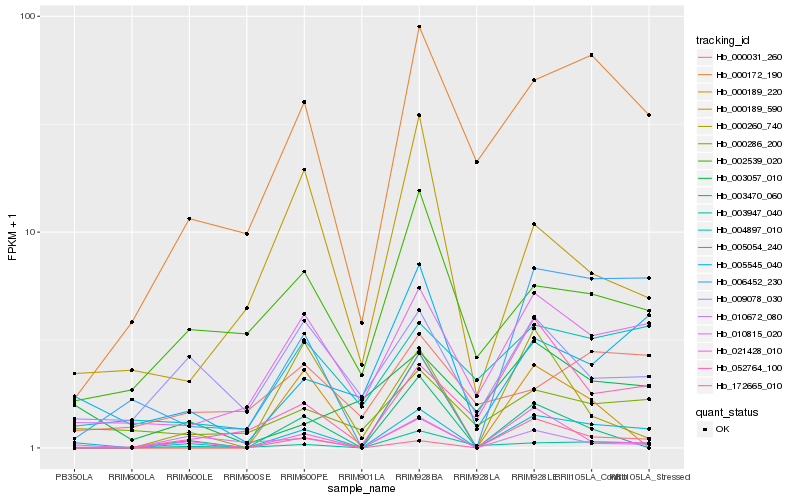
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005054_240 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010815_020 |
0.2584083993 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_005545_040 |
0.2673926744 |
- |
- |
PREDICTED: protein LURP-one-related 14 isoform X2 [Nelumbo nucifera] |
| 4 |
Hb_000172_190 |
0.2725078119 |
- |
- |
PREDICTED: uncharacterized protein LOC105650824 [Jatropha curcas] |
| 5 |
Hb_021428_010 |
0.2763554343 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 6 |
Hb_003947_040 |
0.2793111226 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 7 |
Hb_052764_100 |
0.2928728742 |
- |
- |
PREDICTED: beta-galactosidase 8-like [Jatropha curcas] |
| 8 |
Hb_004897_010 |
0.2981021385 |
- |
- |
- |
| 9 |
Hb_000286_200 |
0.3024188783 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_172665_010 |
0.3045661075 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 11 |
Hb_006452_230 |
0.3089642862 |
- |
- |
PREDICTED: uncharacterized protein LOC105635044 [Jatropha curcas] |
| 12 |
Hb_010672_080 |
0.3212086717 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_00902 [Jatropha curcas] |
| 13 |
Hb_003470_060 |
0.3226368536 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 45 [Jatropha curcas] |
| 14 |
Hb_002539_020 |
0.3275481472 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 15 |
Hb_000189_590 |
0.3279474005 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 16 |
Hb_003057_010 |
0.32880001 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 17 |
Hb_009078_030 |
0.3325049443 |
- |
- |
hypothetical protein PRUPE_ppa016992mg, partial [Prunus persica] |
| 18 |
Hb_000260_740 |
0.3362372281 |
- |
- |
- |
| 19 |
Hb_000189_220 |
0.3368892254 |
- |
- |
- |
| 20 |
Hb_000031_260 |
0.3405084502 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |