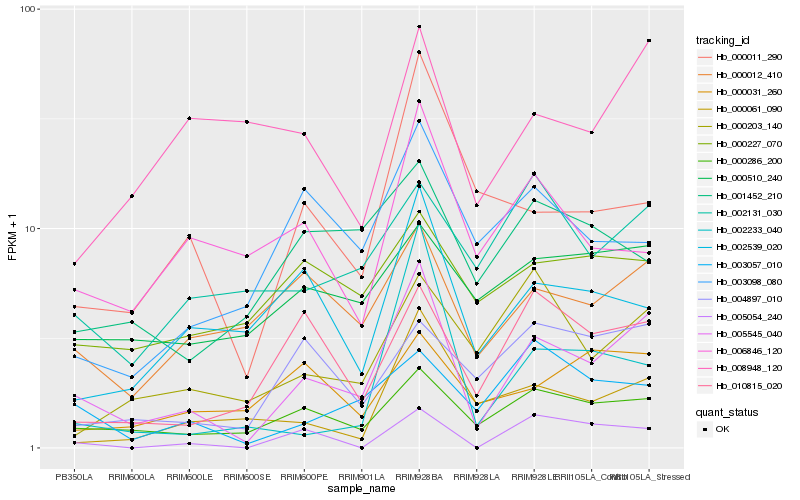
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005545_040 |
0.0 |
- |
- |
PREDICTED: protein LURP-one-related 14 isoform X2 [Nelumbo nucifera] |
| 2 |
Hb_000286_200 |
0.1962863439 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000012_410 |
0.2316092932 |
- |
- |
PREDICTED: histone deacetylase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000011_290 |
0.2368739857 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 5 |
Hb_010815_020 |
0.2390018501 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000061_090 |
0.2409546422 |
- |
- |
hypothetical protein B456_003G0981001, partial [Gossypium raimondii] |
| 7 |
Hb_003057_010 |
0.2437665152 |
- |
- |
hypothetical protein JCGZ_06666 [Jatropha curcas] |
| 8 |
Hb_002539_020 |
0.2493942967 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 9 |
Hb_004897_010 |
0.252594844 |
- |
- |
- |
| 10 |
Hb_000203_140 |
0.2579844264 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 11 |
Hb_003098_080 |
0.2585550368 |
- |
- |
- |
| 12 |
Hb_000510_240 |
0.2586684739 |
desease resistance |
Gene Name: PRK |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_002131_030 |
0.2602595525 |
- |
- |
PREDICTED: pyruvate kinase isozyme G, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000031_260 |
0.2610273997 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 15 |
Hb_002233_040 |
0.2640935628 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 16 |
Hb_000227_070 |
0.2654013712 |
- |
- |
PREDICTED: ATP-dependent (S)-NAD(P)H-hydrate dehydratase isoform X2 [Jatropha curcas] |
| 17 |
Hb_006846_120 |
0.2665444717 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 18 |
Hb_005054_240 |
0.2673926744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_008948_120 |
0.2683227737 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001452_210 |
0.2689038111 |
- |
- |
molybdopterin biosynthesis CNX1 protein [Arabidopsis thaliana] |