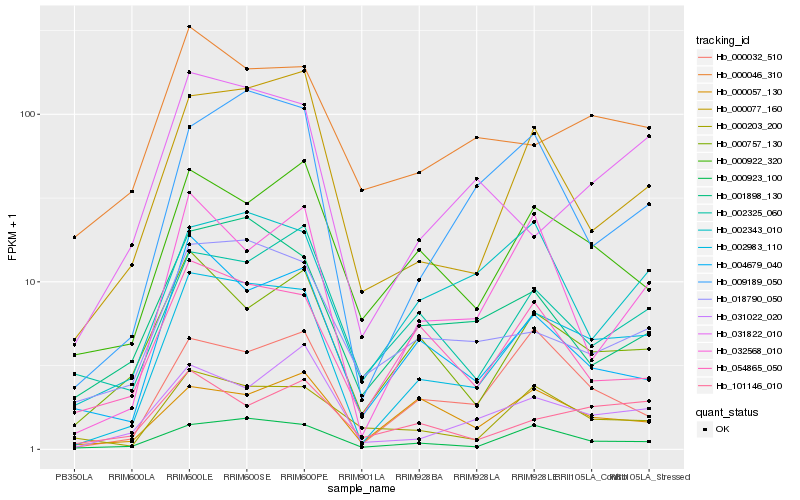
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002983_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634780 [Jatropha curcas] |
| 2 |
Hb_000923_100 |
0.1463426893 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 3 |
Hb_000032_510 |
0.1705070685 |
- |
- |
PREDICTED: SUN domain-containing protein 2 [Jatropha curcas] |
| 4 |
Hb_000077_160 |
0.1718864187 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 5 |
Hb_009189_050 |
0.1739089886 |
- |
- |
PREDICTED: uncharacterized protein LOC105648080 [Jatropha curcas] |
| 6 |
Hb_000203_200 |
0.1789961942 |
- |
- |
Pollen allergen Che a 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000922_320 |
0.1790395443 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 8 |
Hb_000757_130 |
0.179300624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_032568_010 |
0.1801245287 |
- |
- |
PREDICTED: protein phosphatase 2C 56-like [Jatropha curcas] |
| 10 |
Hb_031022_020 |
0.1803123578 |
- |
- |
- |
| 11 |
Hb_002343_010 |
0.1838000688 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_018790_050 |
0.1869993193 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 13 |
Hb_054865_050 |
0.1928125064 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 14 |
Hb_001898_130 |
0.193072204 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_004679_040 |
0.1932537271 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 16 |
Hb_002325_060 |
0.1942106975 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 17 |
Hb_031822_010 |
0.1958479139 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 18 |
Hb_000046_310 |
0.1962413539 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 19 |
Hb_101146_010 |
0.1963301305 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 20 |
Hb_000057_130 |
0.1966310124 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |