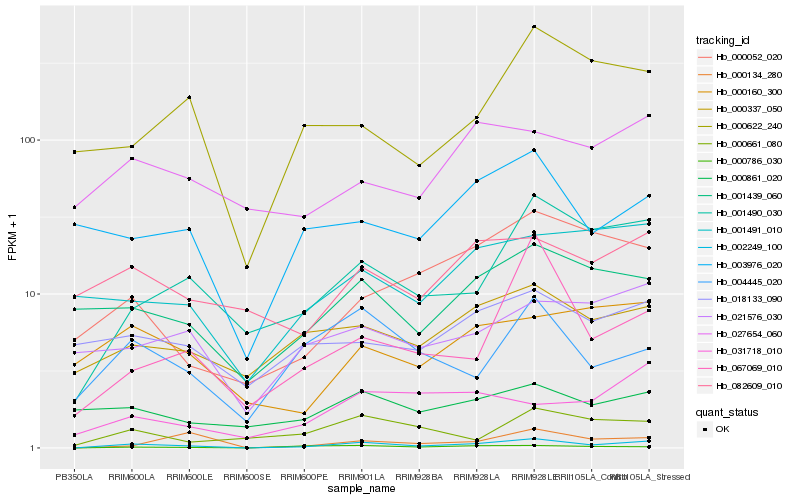
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002249_100 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 2 |
Hb_001490_030 |
0.2212146242 |
- |
- |
PREDICTED: psbP domain-containing protein 6, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000786_030 |
0.242906012 |
- |
- |
PREDICTED: uncharacterized protein LOC104240691 [Nicotiana sylvestris] |
| 4 |
Hb_004445_020 |
0.2472116391 |
- |
- |
hypothetical protein JCGZ_13166 [Jatropha curcas] |
| 5 |
Hb_000337_050 |
0.248288748 |
- |
- |
PREDICTED: chloroplast envelope membrane protein [Jatropha curcas] |
| 6 |
Hb_001439_060 |
0.2484276856 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 7 |
Hb_067069_010 |
0.2553600259 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 8 |
Hb_000052_020 |
0.2594018656 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 9 |
Hb_001491_010 |
0.260179191 |
- |
- |
PREDICTED: uncharacterized protein LOC105649805 [Jatropha curcas] |
| 10 |
Hb_000134_280 |
0.2603592984 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 11 |
Hb_000661_080 |
0.2614255524 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 12 |
Hb_003976_020 |
0.2620859209 |
- |
- |
Regulator of ribonuclease activity A, putative [Ricinus communis] |
| 13 |
Hb_082609_010 |
0.2622853673 |
- |
- |
PREDICTED: uncharacterized protein LOC105644299 [Jatropha curcas] |
| 14 |
Hb_021576_030 |
0.2651569058 |
- |
- |
PREDICTED: uncharacterized protein LOC105637542 [Jatropha curcas] |
| 15 |
Hb_031718_010 |
0.2665407124 |
- |
- |
- |
| 16 |
Hb_000160_300 |
0.2680448769 |
- |
- |
hypothetical protein POPTR_0004s08720g [Populus trichocarpa] |
| 17 |
Hb_000861_020 |
0.2688493023 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 18 |
Hb_018133_090 |
0.2705018267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000622_240 |
0.2711105815 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1A, chloroplastic [Jatropha curcas] |
| 20 |
Hb_027654_060 |
0.2715866637 |
- |
- |
RecName: Full=Probable pyridoxal 5'-phosphate synthase subunit PDX1; Short=PLP synthase subunit PDX1; AltName: Full=Ethylene-inducible protein HEVER [Hevea brasiliensis] |