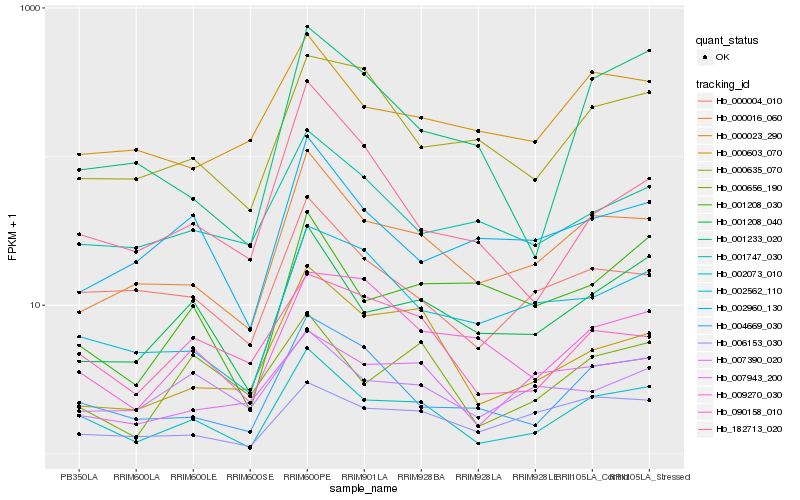
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002073_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000016_060 |
0.1509094909 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 3 |
Hb_001208_040 |
0.1533488008 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |
| 4 |
Hb_004669_030 |
0.1629561213 |
- |
- |
- |
| 5 |
Hb_000603_070 |
0.1771853751 |
- |
- |
- |
| 6 |
Hb_000004_010 |
0.1821496658 |
- |
- |
hypothetical protein Csa_6G120400 [Cucumis sativus] |
| 7 |
Hb_090158_010 |
0.18601483 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |
| 8 |
Hb_000656_190 |
0.1878264701 |
- |
- |
PREDICTED: probable nucleoside diphosphate kinase 5 [Jatropha curcas] |
| 9 |
Hb_001233_020 |
0.1901164537 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 10 |
Hb_006153_030 |
0.1907451971 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 11 |
Hb_002562_110 |
0.1940972398 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 12 |
Hb_001208_030 |
0.1991193135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007943_200 |
0.2018585341 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_000635_070 |
0.204780016 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 15 |
Hb_000023_290 |
0.2051272602 |
- |
- |
steroid binding protein, putative [Ricinus communis] |
| 16 |
Hb_009270_030 |
0.2065166579 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 17 |
Hb_002960_130 |
0.2070037443 |
- |
- |
hypothetical protein B456_008G036100 [Gossypium raimondii] |
| 18 |
Hb_007390_020 |
0.2088741133 |
- |
- |
PREDICTED: protein FMP32, mitochondrial-like [Populus euphratica] |
| 19 |
Hb_001747_030 |
0.20996764 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 20 |
Hb_182713_020 |
0.2112468168 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |