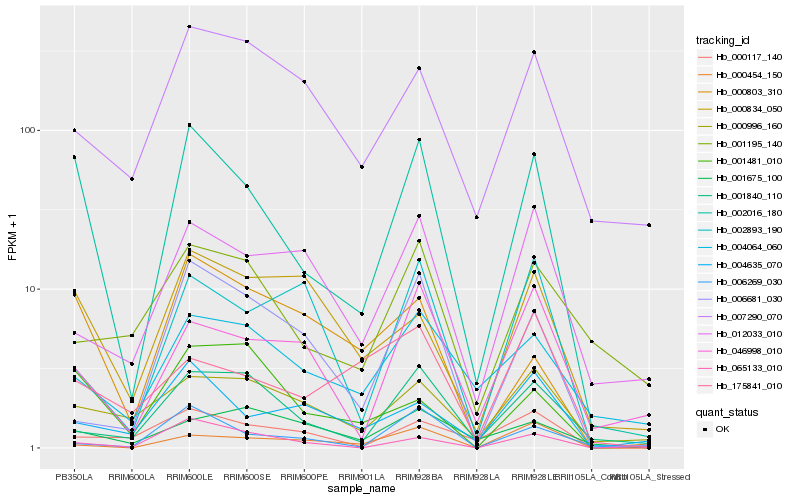
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002016_180 |
0.0 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 2 |
Hb_065133_010 |
0.2582369087 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 3 |
Hb_000803_310 |
0.2588268835 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001840_110 |
0.2628464737 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 5 |
Hb_001481_010 |
0.2630147153 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006269_030 |
0.2633034311 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 7 |
Hb_000996_160 |
0.2636726915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_046998_010 |
0.2654168149 |
desease resistance |
Gene Name: LRR_4 |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 9 |
Hb_004064_060 |
0.2661650976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000117_140 |
0.2681293002 |
- |
- |
hypothetical protein CISIN_1g042884mg, partial [Citrus sinensis] |
| 11 |
Hb_000834_050 |
0.2682247924 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 12 |
Hb_004635_070 |
0.2808880423 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Solanum lycopersicum] |
| 13 |
Hb_007290_070 |
0.2830299871 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 14 |
Hb_001195_140 |
0.2873760638 |
- |
- |
PREDICTED: U-box domain-containing protein 5-like [Jatropha curcas] |
| 15 |
Hb_012033_010 |
0.2886163503 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 16 |
Hb_002893_190 |
0.2886926389 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_006681_030 |
0.2898324199 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 18 |
Hb_000454_150 |
0.2944459107 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 19 |
Hb_175841_010 |
0.2973906736 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 20 |
Hb_001675_100 |
0.2979321175 |
- |
- |
conserved hypothetical protein [Ricinus communis] |