| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
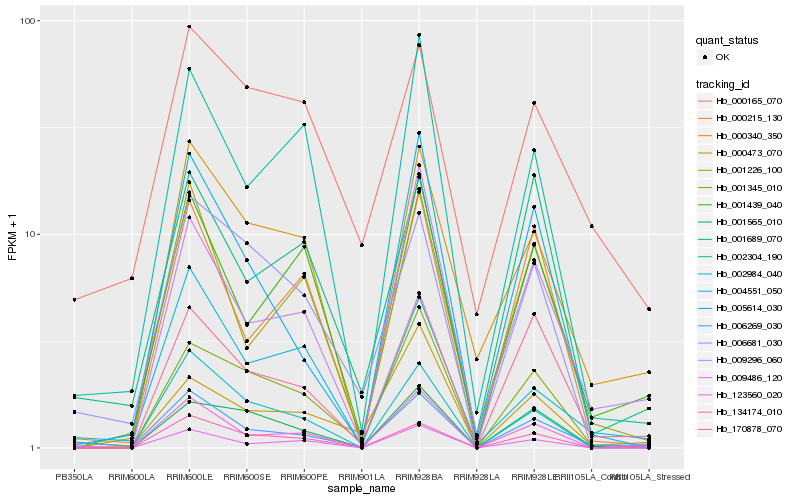
Hb_006269_030 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 2 |
Hb_170878_070 |
0.1679481231 |
- |
- |
hypothetical protein EUGRSUZ_L008133, partial [Eucalyptus grandis] |
| 3 |
Hb_009486_120 |
0.1684933803 |
- |
- |
hypothetical protein PRUPE_ppa004149mg [Prunus persica] |
| 4 |
Hb_134174_010 |
0.175306245 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_006681_030 |
0.1775639982 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_001226_100 |
0.1778700574 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 7 |
Hb_009296_060 |
0.1801739736 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 8 |
Hb_001689_070 |
0.1814014001 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_005614_030 |
0.1816873943 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004551_050 |
0.1819124964 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001439_040 |
0.186837142 |
- |
- |
PREDICTED: chorismate mutase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_123560_020 |
0.1876109289 |
- |
- |
hypothetical protein JCGZ_12464 [Jatropha curcas] |
| 13 |
Hb_000215_130 |
0.1880108671 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 14 |
Hb_001345_010 |
0.1904757436 |
- |
- |
hypothetical protein MIMGU_mgv1a010889mg [Erythranthe guttata] |
| 15 |
Hb_000340_350 |
0.1905409436 |
- |
- |
PREDICTED: sugar transporter ERD6-like 7 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002304_190 |
0.1924732901 |
- |
- |
PREDICTED: 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase [Jatropha curcas] |
| 17 |
Hb_002984_040 |
0.1934991912 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000165_070 |
0.1937509098 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 19 |
Hb_000473_070 |
0.1942916822 |
- |
- |
PREDICTED: amino-acid permease BAT1 homolog [Jatropha curcas] |
| 20 |
Hb_001565_010 |
0.1956384849 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |