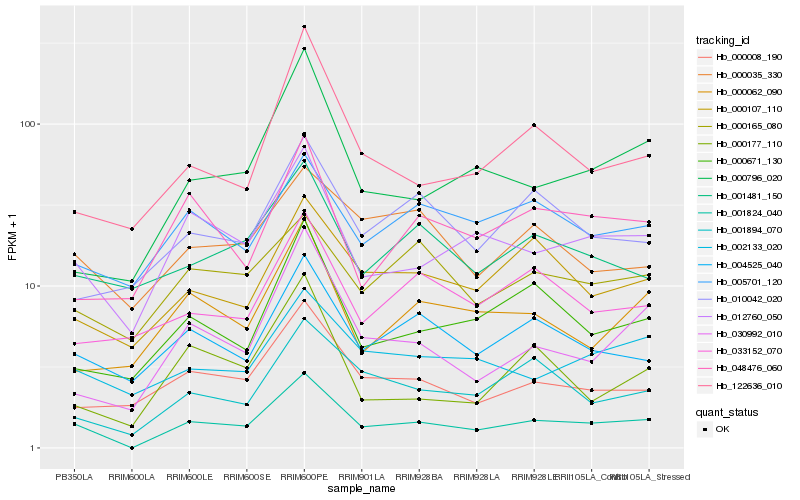
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001824_040 |
0.0 |
- |
- |
hypothetical protein M569_11359, partial [Genlisea aurea] |
| 2 |
Hb_012760_050 |
0.1444285379 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 3 |
Hb_004525_040 |
0.1641334824 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 4 |
Hb_001894_070 |
0.1687164412 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000107_110 |
0.1700498191 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000671_130 |
0.1702007778 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000008_190 |
0.1703752702 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_030992_010 |
0.1725025864 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 9 |
Hb_002133_020 |
0.1752464024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_122636_010 |
0.1797228086 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 11 |
Hb_000796_020 |
0.1811065311 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 12 |
Hb_000062_090 |
0.1832223007 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001481_150 |
0.183518524 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 14 |
Hb_005701_120 |
0.1841895805 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 15 |
Hb_048476_060 |
0.1846538437 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000165_080 |
0.1873425337 |
- |
- |
PREDICTED: multiple inositol polyphosphate phosphatase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_033152_070 |
0.1874052168 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 18 |
Hb_000035_330 |
0.1891574888 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |
| 19 |
Hb_010042_020 |
0.1894653477 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 20 |
Hb_000177_110 |
0.1909285204 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |