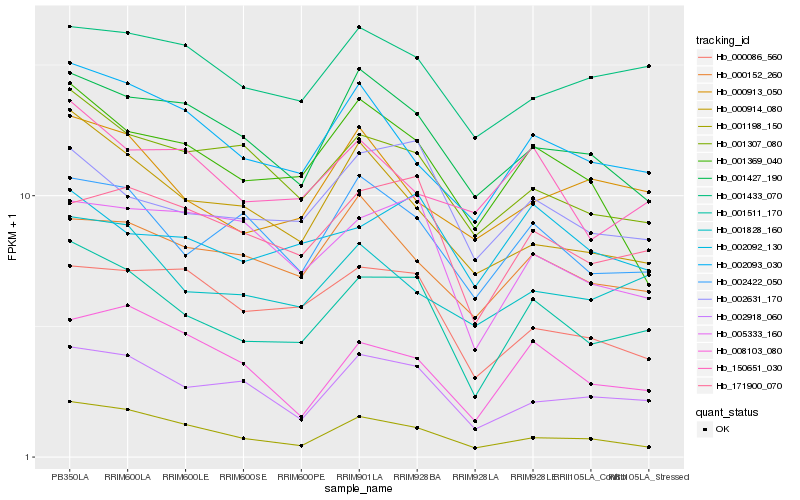
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001511_170 |
0.0 |
- |
- |
PREDICTED: GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 2 |
Hb_002918_060 |
0.0968987116 |
- |
- |
PREDICTED: glyoxysomal processing protease, glyoxysomal isoform X1 [Jatropha curcas] |
| 3 |
Hb_171900_070 |
0.1051158489 |
- |
- |
- |
| 4 |
Hb_002422_050 |
0.1057633574 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002631_170 |
0.1062453198 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 6 |
Hb_002093_030 |
0.1062931291 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 7 |
Hb_001828_160 |
0.1123289879 |
- |
- |
hypothetical protein F775_31105 [Aegilops tauschii] |
| 8 |
Hb_001307_080 |
0.1133511248 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 9 |
Hb_001427_190 |
0.116400181 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 10 |
Hb_000914_080 |
0.1184197674 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_005333_160 |
0.1200495334 |
- |
- |
hypothetical protein B456_009G230200 [Gossypium raimondii] |
| 12 |
Hb_001369_040 |
0.1220800417 |
- |
- |
PREDICTED: probable isoaspartyl peptidase/L-asparaginase 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000152_260 |
0.1223298088 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 14 |
Hb_000913_050 |
0.1225106996 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 15 |
Hb_002092_130 |
0.1232032337 |
- |
- |
PREDICTED: protein CASP-like [Oryza brachyantha] |
| 16 |
Hb_001198_150 |
0.1239744176 |
- |
- |
hypothetical protein POPTR_0003s01280g [Populus trichocarpa] |
| 17 |
Hb_000086_560 |
0.1241889123 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_150651_030 |
0.1247968068 |
- |
- |
PREDICTED: metallophosphoesterase 1-like [Jatropha curcas] |
| 19 |
Hb_001433_070 |
0.1253989954 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 20 |
Hb_008103_080 |
0.1264751766 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |