| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
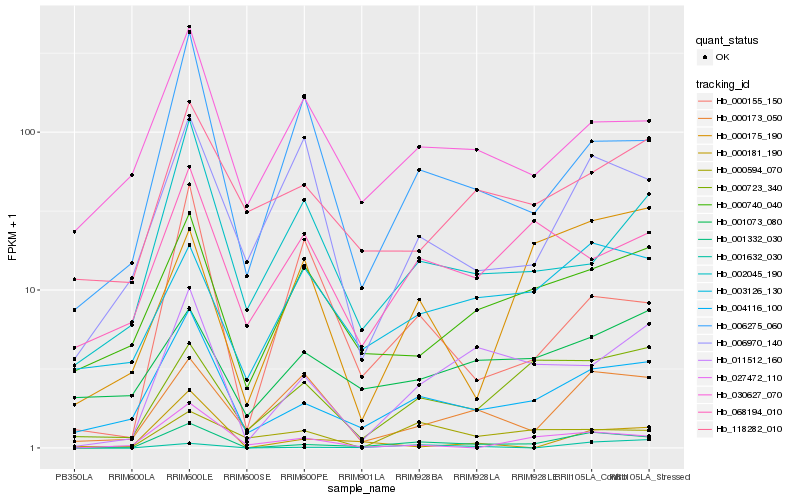
Hb_001332_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_21111 [Jatropha curcas] |
| 2 |
Hb_004116_100 |
0.2024838951 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |
| 3 |
Hb_011512_160 |
0.2305029429 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000723_340 |
0.2658142712 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 5 |
Hb_000740_040 |
0.2819725085 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 6 |
Hb_006275_060 |
0.2823288824 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 7 |
Hb_000173_050 |
0.2828115204 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 8 |
Hb_003126_130 |
0.286494919 |
- |
- |
inositol-1-monophosphatase family protein [Populus trichocarpa] |
| 9 |
Hb_027472_110 |
0.291696591 |
- |
- |
Patellin-4, putative [Ricinus communis] |
| 10 |
Hb_001632_030 |
0.2927007523 |
- |
- |
PREDICTED: chaperone protein ClpB1 [Jatropha curcas] |
| 11 |
Hb_118282_010 |
0.2957075486 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 12 |
Hb_030627_070 |
0.2964743951 |
- |
- |
histone H4 [Zea mays] |
| 13 |
Hb_002045_190 |
0.2996132815 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 14 |
Hb_000181_190 |
0.3008042244 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 15 |
Hb_000155_150 |
0.3020234192 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 16 |
Hb_068194_010 |
0.3044261958 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 17 |
Hb_001073_080 |
0.3046220628 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000175_190 |
0.3050422714 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 19 |
Hb_000594_070 |
0.3053463776 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_006970_140 |
0.3067290709 |
- |
- |
PREDICTED: histone H3.2-like [Solanum lycopersicum] |