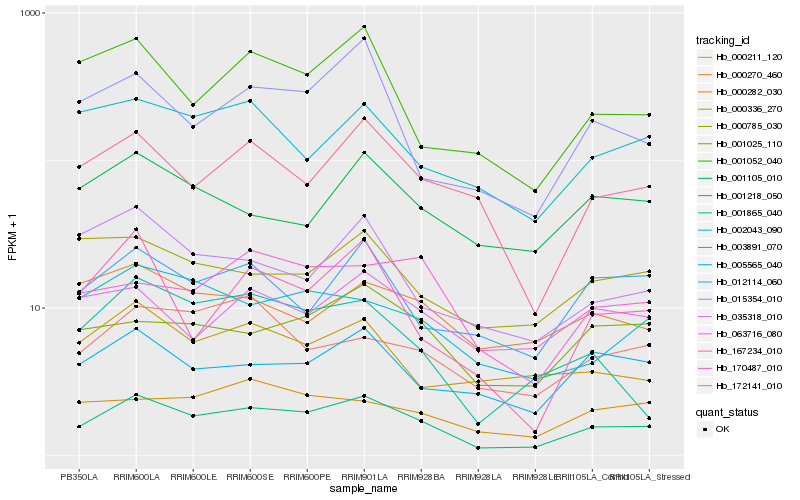
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001218_050 |
0.0 |
- |
- |
Ribosome-binding protein, putative [Ricinus communis] |
| 2 |
Hb_172141_010 |
0.1381931352 |
- |
- |
PREDICTED: uncharacterized protein LOC105641033 [Jatropha curcas] |
| 3 |
Hb_005565_040 |
0.1432089332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001025_110 |
0.1435490941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001052_040 |
0.1466514269 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_012114_060 |
0.1501441261 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 7 |
Hb_003891_070 |
0.1502268162 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 8 |
Hb_015354_010 |
0.1510739817 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 9 |
Hb_002043_090 |
0.1519456803 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 10 |
Hb_000336_270 |
0.1543613417 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_167234_010 |
0.1552020061 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 12 |
Hb_000785_030 |
0.1585572259 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 13 |
Hb_000211_120 |
0.158823081 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 14 |
Hb_000270_460 |
0.1590412405 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 8 [Jatropha curcas] |
| 15 |
Hb_063716_080 |
0.1591882144 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 16 |
Hb_035318_010 |
0.160519291 |
- |
- |
PREDICTED: uncharacterized protein LOC105645250 [Jatropha curcas] |
| 17 |
Hb_001865_040 |
0.1612994845 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 18 |
Hb_000282_030 |
0.1615172705 |
- |
- |
K(+) efflux antiporter 4 [Glycine soja] |
| 19 |
Hb_170487_010 |
0.1617816009 |
- |
- |
- |
| 20 |
Hb_001105_010 |
0.1618664465 |
desease resistance |
Gene Name: AIG1 |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |