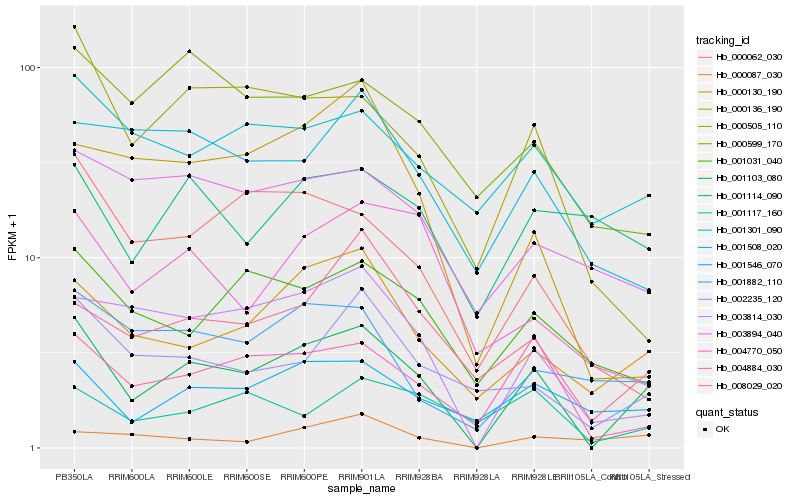
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001103_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_004884_030 |
0.1981820484 |
- |
- |
PREDICTED: ammonium transporter 3 member 3 [Vitis vinifera] |
| 3 |
Hb_000130_190 |
0.2010303867 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 4 |
Hb_003814_030 |
0.2044029391 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_001546_070 |
0.2152411579 |
- |
- |
Uncharacterized protein F383_19872 [Gossypium arboreum] |
| 6 |
Hb_008029_020 |
0.2229540909 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 7 |
Hb_002235_120 |
0.2241424364 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 8 |
Hb_000505_110 |
0.2272796378 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |
| 9 |
Hb_001301_090 |
0.2294996075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001882_110 |
0.2296796543 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001031_040 |
0.2319121827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000062_030 |
0.2407774519 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003894_040 |
0.2409958779 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 14 |
Hb_001117_160 |
0.2426893141 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g10270 [Jatropha curcas] |
| 15 |
Hb_000599_170 |
0.244103543 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2-like isoform X2 [Populus euphratica] |
| 16 |
Hb_001114_090 |
0.2446866187 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 17 |
Hb_000136_190 |
0.2447886392 |
- |
- |
PREDICTED: protein-S-isoprenylcysteine O-methyltransferase A-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000087_030 |
0.2448607303 |
- |
- |
hypothetical protein JCGZ_02243 [Jatropha curcas] |
| 19 |
Hb_001508_020 |
0.2454083586 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 20 |
Hb_004770_050 |
0.2465637446 |
- |
- |
PREDICTED: serine carboxypeptidase-like 35 [Populus euphratica] |