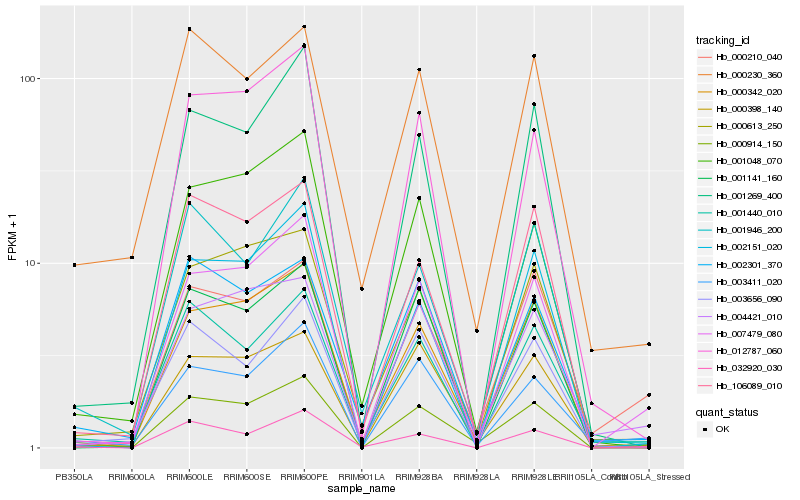
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000914_150 |
0.0 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 2 |
Hb_003411_020 |
0.1234233679 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 3 |
Hb_000398_140 |
0.1244376641 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 4 |
Hb_000342_020 |
0.1286190028 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 5 |
Hb_007479_080 |
0.1300273519 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001440_010 |
0.1315390561 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 7 |
Hb_002301_370 |
0.1392187893 |
- |
- |
hypothetical protein JCGZ_21454 [Jatropha curcas] |
| 8 |
Hb_032920_030 |
0.1400343972 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 9 |
Hb_001946_200 |
0.1416427654 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 10 |
Hb_000230_360 |
0.1423651918 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 11 |
Hb_000613_250 |
0.1429043614 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 12 |
Hb_004421_010 |
0.1429489382 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 13 |
Hb_106089_010 |
0.1431587184 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 14 |
Hb_001269_400 |
0.1451140774 |
- |
- |
cinnamoyl-CoA reductase [Hevea brasiliensis] |
| 15 |
Hb_000210_040 |
0.1476236035 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 16 |
Hb_002151_020 |
0.1485285588 |
- |
- |
PREDICTED: uncharacterized protein LOC105637945 [Jatropha curcas] |
| 17 |
Hb_003656_090 |
0.148735984 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 18 |
Hb_001048_070 |
0.1493872474 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 19 |
Hb_001141_160 |
0.1496420658 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 20 |
Hb_012787_060 |
0.1504227976 |
- |
- |
PREDICTED: glutathione S-transferase U17-like [Prunus mume] |