| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
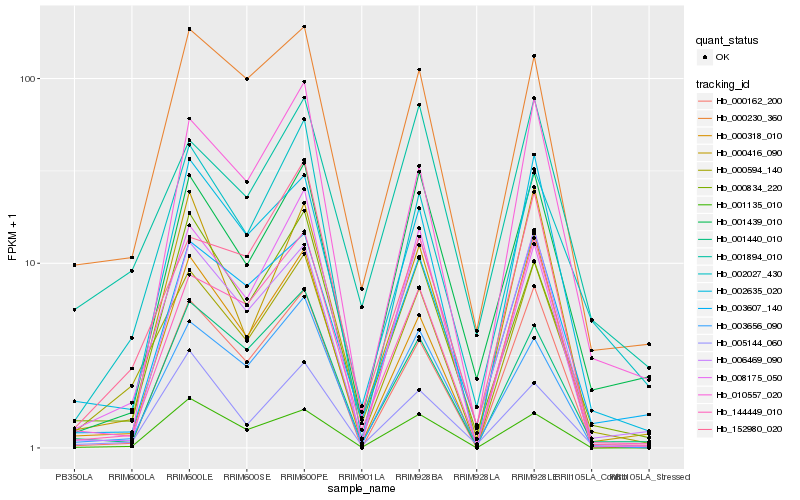
Hb_000594_140 |
0.0 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 2 |
Hb_002635_020 |
0.122243039 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 3 |
Hb_008175_050 |
0.1302877137 |
- |
- |
PREDICTED: formin-like protein 2 [Jatropha curcas] |
| 4 |
Hb_003656_090 |
0.1311072905 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 5 |
Hb_002027_430 |
0.132585062 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 6 |
Hb_144449_010 |
0.1338946459 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 7 |
Hb_000230_360 |
0.1339104673 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 8 |
Hb_003607_140 |
0.1348290399 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 9 |
Hb_000162_200 |
0.1375258296 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001440_010 |
0.139603503 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 1 [Populus euphratica] |
| 11 |
Hb_001894_010 |
0.1405604649 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 12 |
Hb_000834_220 |
0.1419936025 |
- |
- |
PREDICTED: uncharacterized protein LOC105637705 [Jatropha curcas] |
| 13 |
Hb_001135_010 |
0.1436001291 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 14 |
Hb_152980_020 |
0.1441022044 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 15 |
Hb_001439_010 |
0.1443025391 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 5-like [Jatropha curcas] |
| 16 |
Hb_000318_010 |
0.1463621526 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 17 |
Hb_000416_090 |
0.1469892604 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 18 |
Hb_005144_060 |
0.1502361943 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 19 |
Hb_010557_020 |
0.1521202507 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 20 |
Hb_006469_090 |
0.1521665325 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |