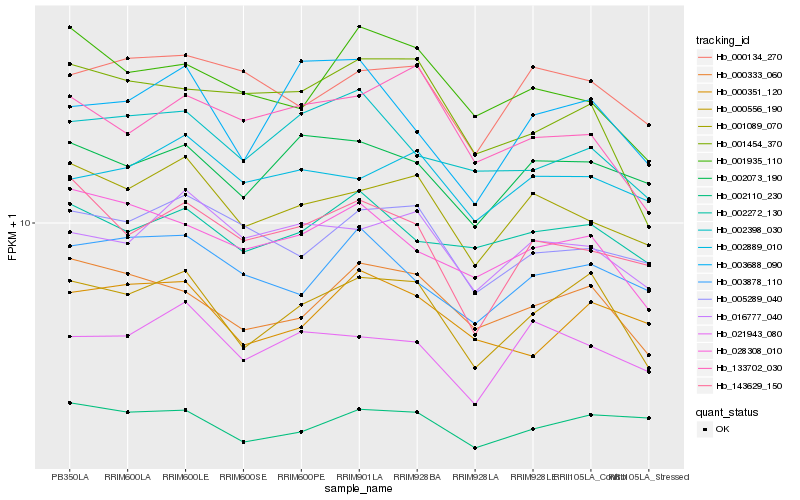
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000556_190 |
0.0 |
- |
- |
PREDICTED: armadillo repeat-containing kinesin-like protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000333_060 |
0.0820286727 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 3 |
Hb_001089_070 |
0.0949676283 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 4 |
Hb_002073_190 |
0.1027608716 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 5 |
Hb_016777_040 |
0.1046283848 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 6 |
Hb_028308_010 |
0.1062099723 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 7 |
Hb_021943_080 |
0.1065387851 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002110_230 |
0.1066867168 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 9 |
Hb_003688_090 |
0.106988868 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001454_370 |
0.1085129706 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 11 |
Hb_002272_130 |
0.108612581 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 12 |
Hb_000351_120 |
0.1086396048 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 13 |
Hb_133702_030 |
0.1122670905 |
- |
- |
PREDICTED: uric acid degradation bifunctional protein TTL isoform X1 [Jatropha curcas] |
| 14 |
Hb_003878_110 |
0.1129341805 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 15 |
Hb_005289_040 |
0.1133160538 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 16 |
Hb_002889_010 |
0.1137554275 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 17 |
Hb_002398_030 |
0.1139098532 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_143629_150 |
0.1140546844 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 19 |
Hb_000134_270 |
0.1141324141 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 20 |
Hb_001935_110 |
0.1152996567 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |