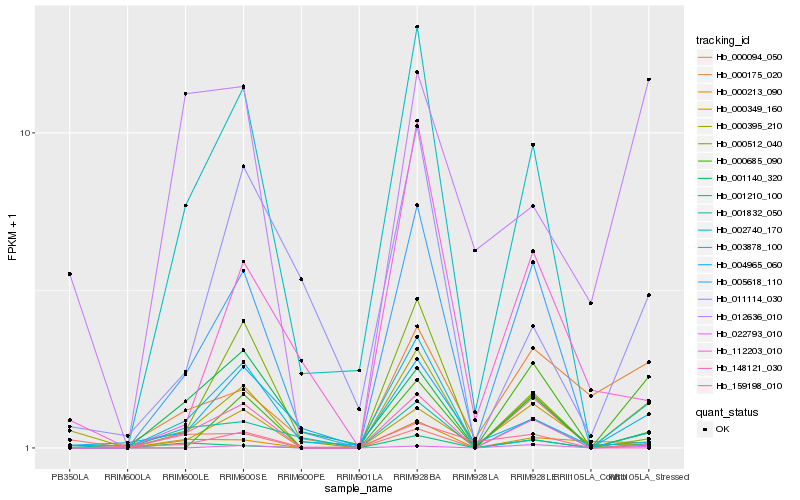
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000395_210 |
0.0 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 2 |
Hb_004965_060 |
0.2776732701 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U4/U6 small nuclear ribonucleoprotein PRP4-like protein [Jatropha curcas] |
| 3 |
Hb_001210_100 |
0.2849383159 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 4 |
Hb_000685_090 |
0.2942596844 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 5 |
Hb_000512_040 |
0.2956716093 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 6 |
Hb_001832_050 |
0.299590726 |
- |
- |
PREDICTED: cytokinin dehydrogenase 3 [Jatropha curcas] |
| 7 |
Hb_022793_010 |
0.3075807152 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 8 |
Hb_000349_160 |
0.3193796656 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Citrus sinensis] |
| 9 |
Hb_000094_050 |
0.3209840418 |
- |
- |
- |
| 10 |
Hb_011114_030 |
0.3248305916 |
- |
- |
PREDICTED: probable carboxylesterase 15 [Jatropha curcas] |
| 11 |
Hb_005618_110 |
0.325062227 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_159198_010 |
0.3252830847 |
- |
- |
hypothetical protein VITISV_033829 [Vitis vinifera] |
| 13 |
Hb_000213_090 |
0.3252925469 |
- |
- |
hypothetical protein OsI_39056 [Oryza sativa Indica Group] |
| 14 |
Hb_003878_100 |
0.3268496808 |
desease resistance |
Gene Name: NB-ARC |
LRR and NB-ARC domains-containing disease resistance protein, putative [Theobroma cacao] |
| 15 |
Hb_002740_170 |
0.3277882585 |
- |
- |
PREDICTED: uncharacterized protein LOC105647357 [Jatropha curcas] |
| 16 |
Hb_112203_010 |
0.3291000879 |
- |
- |
- |
| 17 |
Hb_012636_010 |
0.3301487577 |
- |
- |
- |
| 18 |
Hb_000175_020 |
0.330176655 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001140_320 |
0.3327887199 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |
| 20 |
Hb_148121_030 |
0.3343233283 |
- |
- |
PREDICTED: zinc-finger homeodomain protein 14 [Gossypium raimondii] |