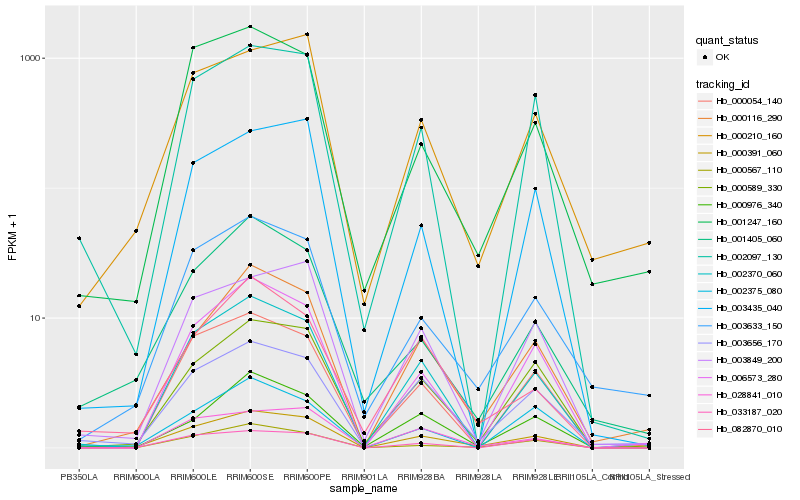
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000391_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104445137 [Eucalyptus grandis] |
| 2 |
Hb_003656_170 |
0.1146246514 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 3 |
Hb_002097_130 |
0.1183032575 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 4 |
Hb_000589_330 |
0.1246164926 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_002370_060 |
0.1301851596 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 6 |
Hb_000116_290 |
0.1343765674 |
- |
- |
PREDICTED: proton-coupled amino acid transporter 3 [Jatropha curcas] |
| 7 |
Hb_003849_200 |
0.1358514587 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_028841_010 |
0.136520785 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase CES101 [Jatropha curcas] |
| 9 |
Hb_033187_020 |
0.1426217237 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 10 |
Hb_000210_160 |
0.1445531075 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 11 |
Hb_003633_150 |
0.1449135466 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 12 |
Hb_082870_010 |
0.1479124395 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 13 |
Hb_000976_340 |
0.1496368713 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000054_140 |
0.1509825732 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1 [Jatropha curcas] |
| 15 |
Hb_006573_280 |
0.1511313351 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 16 |
Hb_001247_160 |
0.1518278833 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 17 |
Hb_000567_110 |
0.1521629583 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 18 |
Hb_003435_040 |
0.1531226883 |
- |
- |
tonoplast intrinsic protein [Hevea brasiliensis] |
| 19 |
Hb_002375_080 |
0.1533761249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001405_060 |
0.1534071697 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 4-like [Jatropha curcas] |