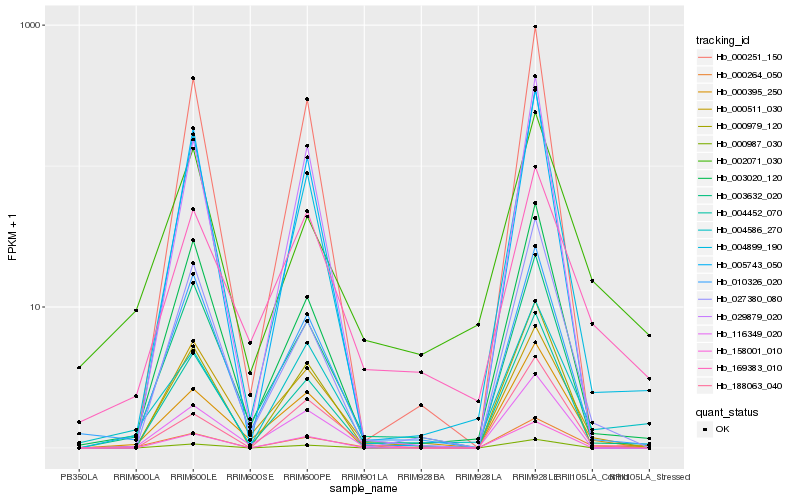
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s04750g [Populus trichocarpa] |
| 2 |
Hb_000395_250 |
0.1576774195 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 3 |
Hb_116349_020 |
0.1585589312 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 4 |
Hb_004899_190 |
0.1663787315 |
- |
- |
RecName: Full=Peroxiredoxin Q, chloroplastic; AltName: Full=Thioredoxin reductase; Flags: Precursor [Populus x jackii] |
| 5 |
Hb_003632_020 |
0.1665240133 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 6 |
Hb_188063_040 |
0.1667810161 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_004586_270 |
0.1686562376 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_010326_020 |
0.1691401115 |
- |
- |
PREDICTED: thioredoxin-like 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000979_120 |
0.1709709091 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 10 |
Hb_003020_120 |
0.1726745511 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 11 |
Hb_000987_030 |
0.1749807805 |
transcription factor |
TF Family: BES1 |
hypothetical protein POPTR_0016s13350g [Populus trichocarpa] |
| 12 |
Hb_004452_070 |
0.175616717 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000511_030 |
0.1757356322 |
- |
- |
PREDICTED: uncharacterized protein LOC105636463 [Jatropha curcas] |
| 14 |
Hb_000251_150 |
0.1757686542 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_005743_050 |
0.1770184068 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_029879_020 |
0.1776741787 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 17 |
Hb_002071_030 |
0.1788514055 |
- |
- |
PREDICTED: ACT domain-containing protein ACR11-like isoform X1 [Gossypium raimondii] |
| 18 |
Hb_169383_010 |
0.1789558817 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 19 |
Hb_027380_080 |
0.17904009 |
- |
- |
hypothetical protein JCGZ_04649 [Jatropha curcas] |
| 20 |
Hb_158001_010 |
0.1790433176 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |