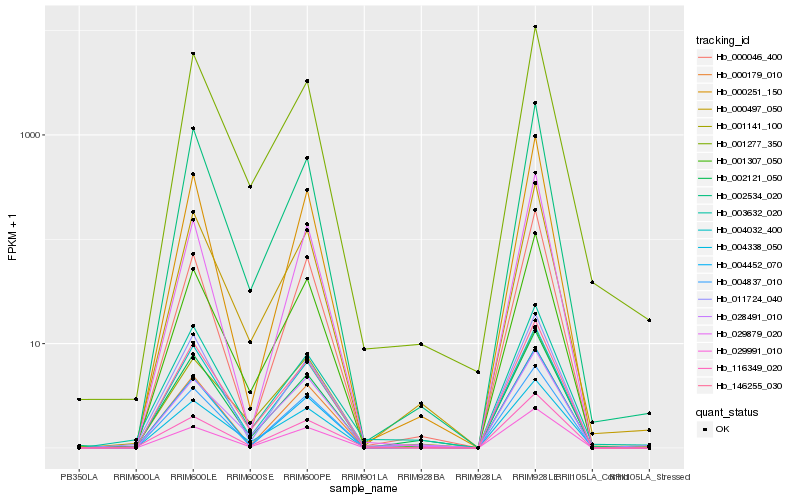
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116349_020 |
0.0 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 2 |
Hb_001307_050 |
0.0711208886 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |
| 3 |
Hb_000179_010 |
0.0842239581 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 4 |
Hb_029991_010 |
0.0873828339 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 5 |
Hb_004837_010 |
0.0886351467 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 6 |
Hb_000497_050 |
0.090037876 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_146255_030 |
0.0929793519 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 8 |
Hb_000046_400 |
0.0933309171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004452_070 |
0.0949679987 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_011724_040 |
0.0968079801 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 11 |
Hb_000251_150 |
0.0983211738 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_028491_010 |
0.0985895026 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 13 |
Hb_029879_020 |
0.0987426476 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |
| 14 |
Hb_001277_350 |
0.0995287034 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 15 |
Hb_004338_050 |
0.0997612018 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 16 |
Hb_003632_020 |
0.1001233312 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 17 |
Hb_001141_100 |
0.1003927159 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 18 |
Hb_004032_400 |
0.1012119058 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 19 |
Hb_002534_020 |
0.1016723576 |
- |
- |
PREDICTED: chlorophyll a-b binding protein 4, chloroplastic [Gossypium raimondii] |
| 20 |
Hb_002121_050 |
0.1018491683 |
- |
- |
hypothetical protein EUGRSUZ_D02586 [Eucalyptus grandis] |