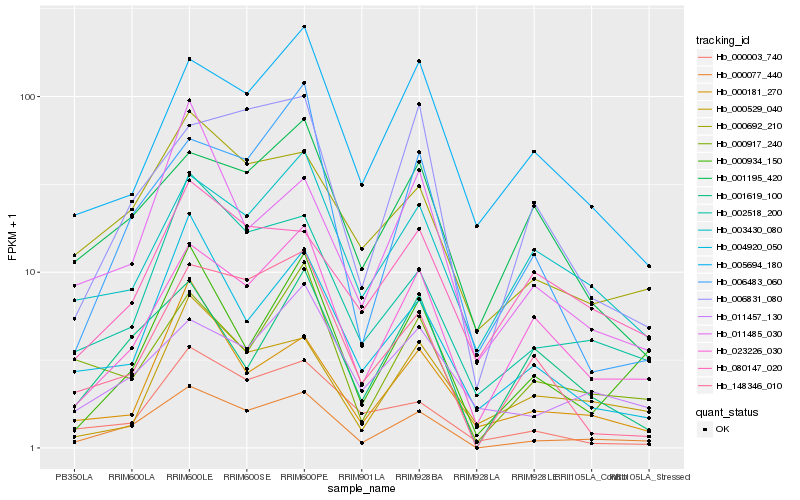
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_440 |
0.0 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 2 |
Hb_002518_200 |
0.1442744641 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |
| 3 |
Hb_000692_210 |
0.1514449813 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_023226_030 |
0.1528575838 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 5 |
Hb_148346_010 |
0.1732247176 |
- |
- |
- |
| 6 |
Hb_000934_150 |
0.1803998266 |
- |
- |
hypothetical protein POPTR_0005s21000g [Populus trichocarpa] |
| 7 |
Hb_004920_050 |
0.1839629575 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 8 |
Hb_000917_240 |
0.1848811999 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 9 |
Hb_006831_080 |
0.1852618544 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 10 |
Hb_000003_740 |
0.1860677367 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 11 |
Hb_011457_130 |
0.1878054959 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 12 |
Hb_003430_080 |
0.1883598105 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_006483_060 |
0.1887286255 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |
| 14 |
Hb_001619_100 |
0.1915815436 |
- |
- |
PREDICTED: uncharacterized protein LOC105630593 [Jatropha curcas] |
| 15 |
Hb_005694_180 |
0.1974018061 |
- |
- |
PREDICTED: cytochrome b5 [Jatropha curcas] |
| 16 |
Hb_001195_420 |
0.1978193667 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 17 |
Hb_000181_270 |
0.1985613058 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000529_040 |
0.2016043569 |
- |
- |
PREDICTED: uncharacterized protein LOC105641671 [Jatropha curcas] |
| 19 |
Hb_011485_030 |
0.2020947925 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 20 |
Hb_080147_020 |
0.2043173171 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |