| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
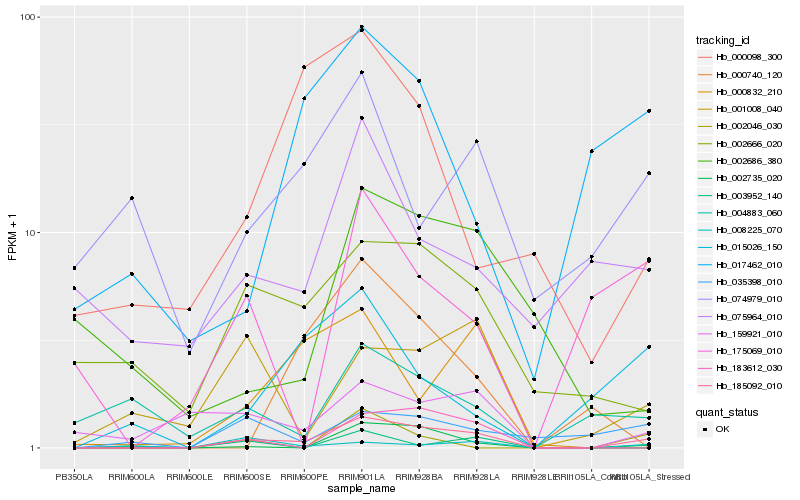
Hb_185092_010 |
0.0 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_002735_020 |
0.3092403682 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_183612_030 |
0.3141779354 |
- |
- |
PREDICTED: uncharacterized protein LOC105632256 [Jatropha curcas] |
| 4 |
Hb_159921_010 |
0.3376937442 |
- |
- |
- |
| 5 |
Hb_002666_020 |
0.3397237772 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 6 |
Hb_035398_010 |
0.3415596144 |
- |
- |
hypothetical protein JCGZ_13029 [Jatropha curcas] |
| 7 |
Hb_001008_040 |
0.3506794686 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 8 |
Hb_015026_150 |
0.364946492 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 9 |
Hb_008225_070 |
0.3662748547 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 10 |
Hb_017462_010 |
0.3693270773 |
- |
- |
- |
| 11 |
Hb_000832_210 |
0.3700219361 |
- |
- |
PREDICTED: WAT1-related protein At2g39510 [Jatropha curcas] |
| 12 |
Hb_175069_010 |
0.3715085902 |
- |
- |
hypothetical protein POPTR_0002s210401g, partial [Populus trichocarpa] |
| 13 |
Hb_075964_010 |
0.3778167747 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 14 |
Hb_000740_120 |
0.3778184077 |
- |
- |
- |
| 15 |
Hb_003952_140 |
0.3795511163 |
- |
- |
hypothetical protein POPTR_0001s24960g [Populus trichocarpa] |
| 16 |
Hb_002046_030 |
0.3795627484 |
- |
- |
PREDICTED: uncharacterized protein LOC104788746 [Camelina sativa] |
| 17 |
Hb_002686_380 |
0.3809236998 |
- |
- |
- |
| 18 |
Hb_000098_300 |
0.381030648 |
- |
- |
- |
| 19 |
Hb_004883_060 |
0.3811230511 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 20 |
Hb_074979_010 |
0.3813331846 |
- |
- |
hypothetical protein AALP_AA2G213100 [Arabis alpina] |