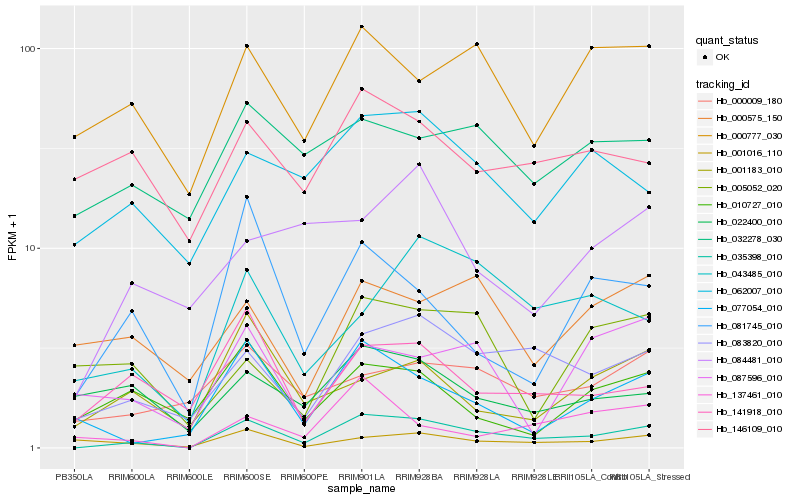
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035398_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_13029 [Jatropha curcas] |
| 2 |
Hb_083820_010 |
0.2074381201 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 3 |
Hb_077054_010 |
0.2294033254 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 4 |
Hb_043485_010 |
0.2517740892 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_141918_010 |
0.2552461168 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 6 |
Hb_001016_110 |
0.2570562923 |
desease resistance |
Gene Name: Lectin_legB |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 7 |
Hb_081745_010 |
0.2579586511 |
- |
- |
hypothetical protein CISIN_1g030953mg [Citrus sinensis] |
| 8 |
Hb_062007_010 |
0.2584449569 |
- |
- |
PREDICTED: argininosuccinate lyase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000009_180 |
0.2603138285 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_137461_010 |
0.2604626136 |
- |
- |
hypothetical protein POPTR_0011s03680g [Populus trichocarpa] |
| 11 |
Hb_000777_030 |
0.2630651535 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 12 |
Hb_001183_010 |
0.2716126587 |
- |
- |
- |
| 13 |
Hb_022400_010 |
0.2721673396 |
- |
- |
Nitrilase/cyanide hydratase and apolipoprotein N-acyltransferase family protein [Theobroma cacao] |
| 14 |
Hb_084481_010 |
0.2734796776 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 15 |
Hb_000575_150 |
0.2761694922 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 16 |
Hb_010727_010 |
0.2767384614 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 17 |
Hb_032278_030 |
0.2781840331 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_087596_010 |
0.2796613383 |
- |
- |
PREDICTED: disease resistance protein RPS4-like [Jatropha curcas] |
| 19 |
Hb_146109_010 |
0.2826082401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005052_020 |
0.2826168225 |
- |
- |
- |