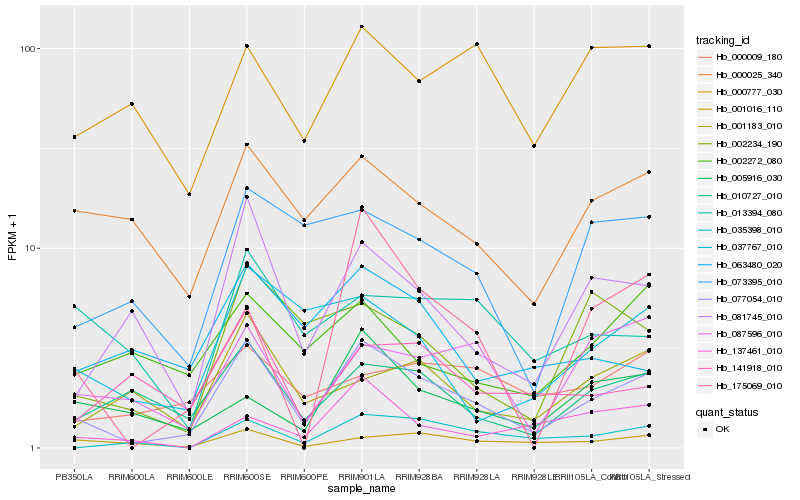
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_077054_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 2 |
Hb_010727_010 |
0.2150324344 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 3 |
Hb_081745_010 |
0.215288863 |
- |
- |
hypothetical protein CISIN_1g030953mg [Citrus sinensis] |
| 4 |
Hb_001016_110 |
0.2266660262 |
desease resistance |
Gene Name: Lectin_legB |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 5 |
Hb_000025_340 |
0.2267184184 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 6 |
Hb_087596_010 |
0.2276785857 |
- |
- |
PREDICTED: disease resistance protein RPS4-like [Jatropha curcas] |
| 7 |
Hb_073395_010 |
0.2290237648 |
- |
- |
- |
| 8 |
Hb_035398_010 |
0.2294033254 |
- |
- |
hypothetical protein JCGZ_13029 [Jatropha curcas] |
| 9 |
Hb_037767_010 |
0.2326177402 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_002234_190 |
0.233404256 |
- |
- |
PREDICTED: putative non-specific lipid-transfer protein 14 [Jatropha curcas] |
| 11 |
Hb_005916_030 |
0.2399791402 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 12 |
Hb_001183_010 |
0.2454634096 |
- |
- |
- |
| 13 |
Hb_000777_030 |
0.2471841256 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 14 |
Hb_175069_010 |
0.2488580638 |
- |
- |
hypothetical protein POPTR_0002s210401g, partial [Populus trichocarpa] |
| 15 |
Hb_000009_180 |
0.2492037589 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 16 |
Hb_002272_080 |
0.252828162 |
- |
- |
- |
| 17 |
Hb_137461_010 |
0.2531447986 |
- |
- |
hypothetical protein POPTR_0011s03680g [Populus trichocarpa] |
| 18 |
Hb_141918_010 |
0.2533808476 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 19 |
Hb_063480_020 |
0.2561013179 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_013394_080 |
0.2561654269 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1D isoform X2 [Jatropha curcas] |