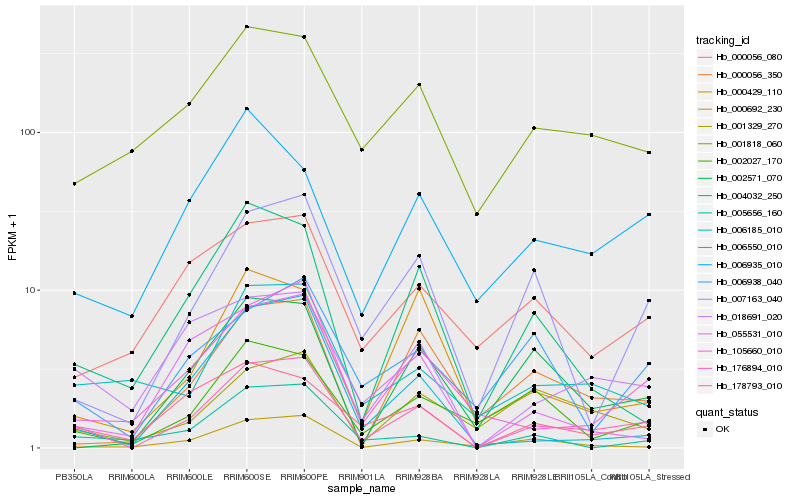
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_176894_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_018691_020 |
0.1903160932 |
- |
- |
- |
| 3 |
Hb_178793_010 |
0.1970983969 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L3 [Nelumbo nucifera] |
| 4 |
Hb_105660_010 |
0.1996951119 |
- |
- |
PREDICTED: potassium channel AKT6-like [Jatropha curcas] |
| 5 |
Hb_007163_040 |
0.2025503605 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Vitis vinifera] |
| 6 |
Hb_002027_170 |
0.2153117184 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 7 |
Hb_006185_010 |
0.2180073772 |
- |
- |
hypothetical protein L484_013453 [Morus notabilis] |
| 8 |
Hb_006550_010 |
0.2181013412 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 9 |
Hb_000692_230 |
0.218308784 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 10 |
Hb_004032_250 |
0.2187522108 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005656_160 |
0.221023806 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |
| 12 |
Hb_001818_060 |
0.2230942383 |
- |
- |
PREDICTED: cysteine proteinase inhibitor 5-like [Jatropha curcas] |
| 13 |
Hb_000056_350 |
0.2237941659 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |
| 14 |
Hb_000429_110 |
0.227432031 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2-like [Populus euphratica] |
| 15 |
Hb_055531_010 |
0.2290736456 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Pyrus x bretschneideri] |
| 16 |
Hb_006938_040 |
0.2295570216 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 17 |
Hb_002571_070 |
0.2307731567 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 18 |
Hb_000056_080 |
0.2311880942 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001329_270 |
0.2323912411 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 20 |
Hb_006935_010 |
0.232699567 |
- |
- |
PREDICTED: inositol transporter 1 isoform X1 [Gossypium raimondii] |