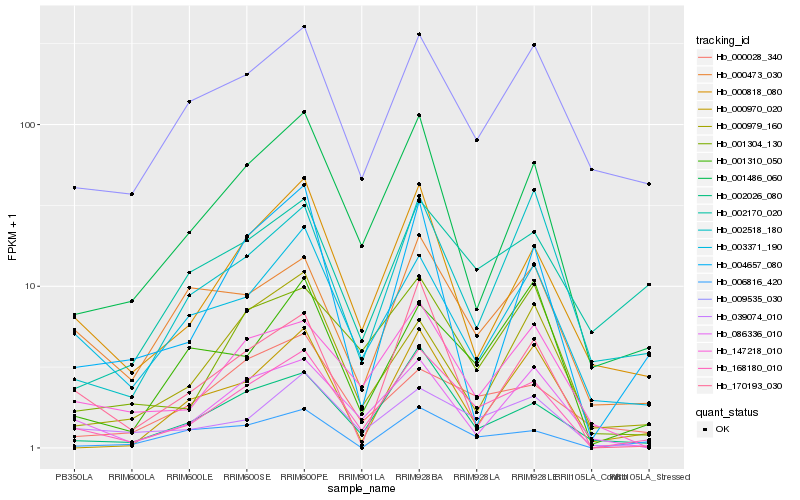
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_168180_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001486_060 |
0.149404258 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 3 |
Hb_000818_080 |
0.1508706297 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 4 |
Hb_002026_080 |
0.1542735219 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 5 |
Hb_039074_010 |
0.1588058864 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_001304_130 |
0.1691263332 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like transcription factor PAT1 [Jatropha curcas] |
| 7 |
Hb_003371_190 |
0.1728249499 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 8 |
Hb_147218_010 |
0.1761671348 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_086336_010 |
0.1836964556 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 10 |
Hb_009535_030 |
0.1851653923 |
- |
- |
CP2 [Hevea brasiliensis] |
| 11 |
Hb_000473_030 |
0.1926799649 |
- |
- |
hypothetical protein JCGZ_19297 [Jatropha curcas] |
| 12 |
Hb_001310_050 |
0.1948722808 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 13 |
Hb_006816_420 |
0.1957586621 |
- |
- |
hypothetical protein JCGZ_23261 [Jatropha curcas] |
| 14 |
Hb_170193_030 |
0.1965537364 |
- |
- |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 15 |
Hb_002518_180 |
0.1971516432 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 16 |
Hb_000028_340 |
0.2012888063 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 17 |
Hb_004657_080 |
0.202644894 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 18 |
Hb_000970_020 |
0.2036899223 |
- |
- |
hypothetical protein POPTR_0005s19050g [Populus trichocarpa] |
| 19 |
Hb_000979_160 |
0.2045546774 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 20 |
Hb_002170_020 |
0.2063929462 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |