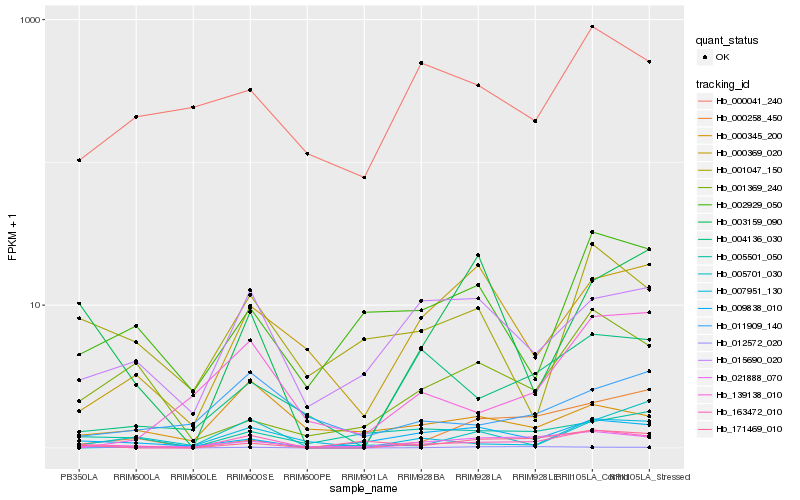
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163472_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_021888_070 |
0.2475994501 |
- |
- |
Putative CCR4-associated factor 1 like 11 [Glycine soja] |
| 3 |
Hb_009838_010 |
0.2758200879 |
- |
- |
- |
| 4 |
Hb_015690_020 |
0.2787030301 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 5 |
Hb_004136_030 |
0.2813604846 |
- |
- |
- |
| 6 |
Hb_000258_450 |
0.3076341036 |
- |
- |
- |
| 7 |
Hb_005501_050 |
0.3077509509 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_139138_010 |
0.3092498535 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 9 |
Hb_011909_140 |
0.3123456815 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 10 |
Hb_007951_130 |
0.3123553355 |
desease resistance |
Gene Name: UBN2_3 |
JHL25H03.6 [Jatropha curcas] |
| 11 |
Hb_003159_090 |
0.3131840386 |
- |
- |
- |
| 12 |
Hb_171469_010 |
0.3134145519 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 13 |
Hb_000369_020 |
0.3143901448 |
- |
- |
PREDICTED: probable fructokinase-7 [Jatropha curcas] |
| 14 |
Hb_001047_150 |
0.3147785433 |
- |
- |
PREDICTED: NF-kappa-B-activating protein [Jatropha curcas] |
| 15 |
Hb_000345_200 |
0.3163495085 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 16 |
Hb_002929_050 |
0.3165676629 |
- |
- |
PREDICTED: polyadenylate-binding protein 6 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005701_030 |
0.3169400891 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001369_240 |
0.3230319727 |
- |
- |
- |
| 19 |
Hb_000041_240 |
0.3232014577 |
- |
- |
glutaredoxin [Hevea brasiliensis] |
| 20 |
Hb_012572_020 |
0.3246935138 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |