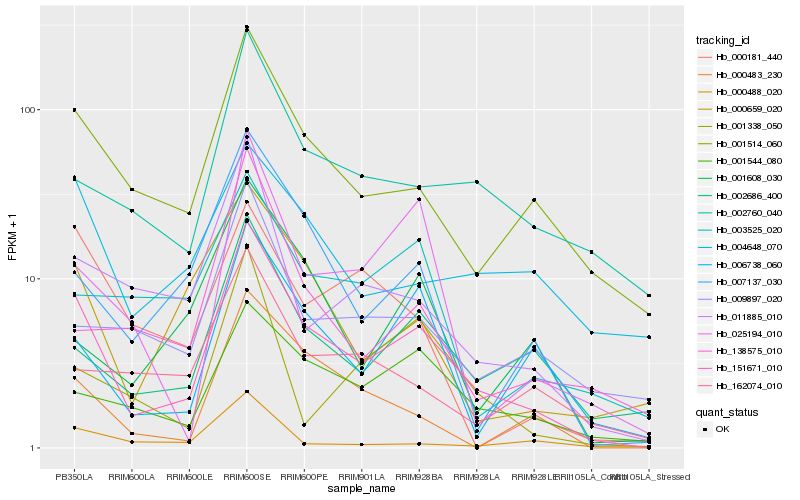
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_151671_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s04441g [Populus trichocarpa] |
| 2 |
Hb_001514_060 |
0.1731522542 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 3 |
Hb_002686_400 |
0.199243533 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 4 |
Hb_007137_030 |
0.2085747632 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.3-like [Jatropha curcas] |
| 5 |
Hb_000659_020 |
0.2096394822 |
- |
- |
PREDICTED: putative L-type lectin-domain containing receptor kinase II.2 [Jatropha curcas] |
| 6 |
Hb_006738_060 |
0.2148929625 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_001544_080 |
0.2154768482 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 8 |
Hb_011885_010 |
0.2160927727 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 9 |
Hb_002760_040 |
0.2181090895 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 10 |
Hb_000483_230 |
0.2236276779 |
- |
- |
PREDICTED: cationic amino acid transporter 6, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_025194_010 |
0.2281385288 |
- |
- |
PREDICTED: uncharacterized protein LOC105644857 [Jatropha curcas] |
| 12 |
Hb_009897_020 |
0.2290113936 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 13 |
Hb_004648_070 |
0.2369100722 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_001608_030 |
0.2378902023 |
- |
- |
Protein NSP-INTERACTING KINASE 1 [Glycine soja] |
| 15 |
Hb_138575_010 |
0.2391849703 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_000488_020 |
0.2422623147 |
- |
- |
PREDICTED: UDP-glucose iridoid glucosyltransferase-like [Jatropha curcas] |
| 17 |
Hb_003525_020 |
0.2432674072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_162074_010 |
0.2452580871 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 19 |
Hb_001338_050 |
0.2468454478 |
- |
- |
hypothetical protein POPTR_0016s05400g [Populus trichocarpa] |
| 20 |
Hb_000181_440 |
0.2482435919 |
- |
- |
- |