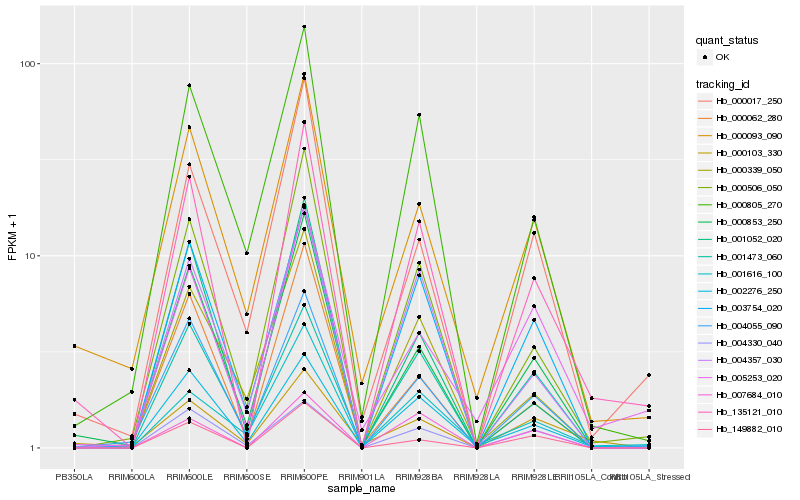
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135121_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635544 [Jatropha curcas] |
| 2 |
Hb_000506_050 |
0.1374107546 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |
| 3 |
Hb_000853_250 |
0.1396102379 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 4 |
Hb_004055_090 |
0.14428377 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 [Jatropha curcas] |
| 5 |
Hb_002276_250 |
0.1451364408 |
- |
- |
PREDICTED: WD repeat and HMG-box DNA-binding protein 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003754_020 |
0.1524288277 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_149882_010 |
0.1583687296 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Populus euphratica] |
| 8 |
Hb_004357_030 |
0.1635578054 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 9 |
Hb_000805_270 |
0.1641804867 |
- |
- |
PREDICTED: expansin-A4 [Gossypium raimondii] |
| 10 |
Hb_000093_090 |
0.1662241708 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 11 |
Hb_000017_250 |
0.1682628956 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 12 |
Hb_007684_010 |
0.1693209824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005253_020 |
0.1730694376 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 14 |
Hb_000062_280 |
0.1745605906 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 15 |
Hb_004330_040 |
0.1787342213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000103_330 |
0.1798068897 |
- |
- |
PREDICTED: meiotic recombination protein DMC1 homolog isoform X1 [Glycine max] |
| 17 |
Hb_001473_060 |
0.1804147005 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |
| 18 |
Hb_001616_100 |
0.1813148638 |
- |
- |
PREDICTED: cell division control protein 45 homolog [Jatropha curcas] |
| 19 |
Hb_000339_050 |
0.1813845845 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 2 [Jatropha curcas] |
| 20 |
Hb_001052_020 |
0.1822127797 |
- |
- |
hypothetical protein JCGZ_26487 [Jatropha curcas] |