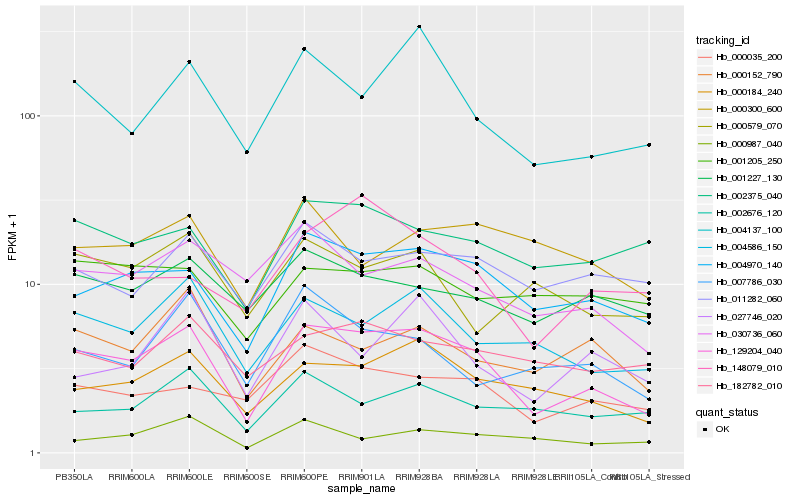
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129204_040 |
0.0 |
- |
- |
PREDICTED: meiotic nuclear division protein 1 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_004970_140 |
0.1306439873 |
- |
- |
PREDICTED: uncharacterized protein LOC105633635 [Jatropha curcas] |
| 3 |
Hb_000184_240 |
0.1369636378 |
- |
- |
PREDICTED: uncharacterized protein LOC105647985 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004586_150 |
0.1468661994 |
- |
- |
PREDICTED: uncharacterized protein LOC105630688 [Jatropha curcas] |
| 5 |
Hb_004137_100 |
0.1549919473 |
- |
- |
PREDICTED: alcohol dehydrogenase 1 [Jatropha curcas] |
| 6 |
Hb_000035_200 |
0.1627686739 |
- |
- |
PREDICTED: phospholipase D Z-like [Jatropha curcas] |
| 7 |
Hb_030736_060 |
0.1671352277 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 8 |
Hb_000152_790 |
0.1671692194 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 9 |
Hb_002375_040 |
0.1700319113 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 10 |
Hb_002676_120 |
0.1718447817 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 11 |
Hb_001227_130 |
0.1749141867 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 12 |
Hb_011282_060 |
0.1770072289 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 13 |
Hb_182782_010 |
0.1785709484 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000987_040 |
0.1787982473 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 2 homolog B isoform X2 [Jatropha curcas] |
| 15 |
Hb_148079_010 |
0.1793220427 |
- |
- |
PREDICTED: WD repeat-containing protein 82 [Jatropha curcas] |
| 16 |
Hb_007786_030 |
0.182624848 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 17 |
Hb_027746_020 |
0.184790735 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit B [Jatropha curcas] |
| 18 |
Hb_000579_070 |
0.1878163335 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 19 |
Hb_001205_250 |
0.1884589244 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 20 |
Hb_000300_600 |
0.1885203507 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |