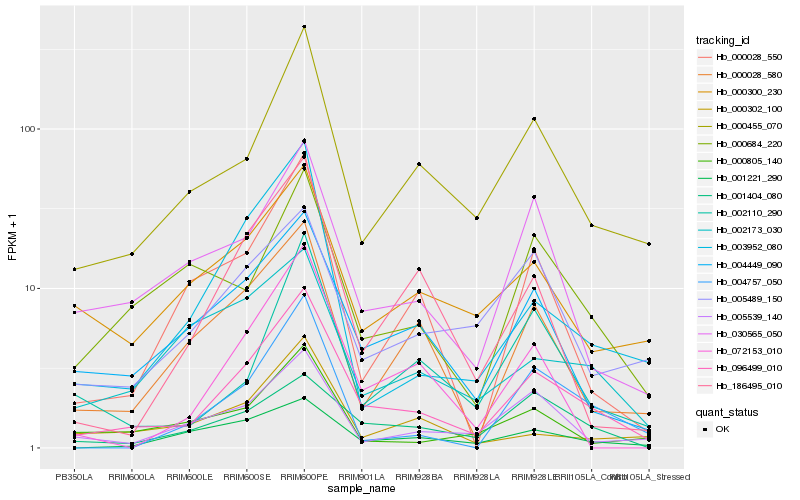
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096499_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 2 |
Hb_000455_070 |
0.160427108 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 3 |
Hb_004449_090 |
0.1725395805 |
- |
- |
PREDICTED: CO(2)-response secreted protease-like [Jatropha curcas] |
| 4 |
Hb_030565_050 |
0.191272574 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 5 |
Hb_000028_580 |
0.1978417674 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 6 |
Hb_003952_080 |
0.2004022267 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_002110_290 |
0.2020537301 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_000300_230 |
0.202142402 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 9 |
Hb_005539_140 |
0.2024066538 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 10 |
Hb_001221_290 |
0.2054059972 |
- |
- |
hypothetical protein RCOM_1506010 [Ricinus communis] |
| 11 |
Hb_186495_010 |
0.2055925014 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 12 |
Hb_000028_550 |
0.2067900662 |
- |
- |
unknown [Medicago truncatula] |
| 13 |
Hb_072153_010 |
0.2083982395 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001404_080 |
0.2092205266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005489_150 |
0.2103827105 |
- |
- |
hypothetical protein JCGZ_20759 [Jatropha curcas] |
| 16 |
Hb_000684_220 |
0.210621284 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_004757_050 |
0.2130740857 |
- |
- |
- |
| 18 |
Hb_000805_140 |
0.2139612146 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 19 |
Hb_002173_030 |
0.2157906547 |
- |
- |
hypothetical protein JCGZ_12466 [Jatropha curcas] |
| 20 |
Hb_000302_100 |
0.2165372833 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |