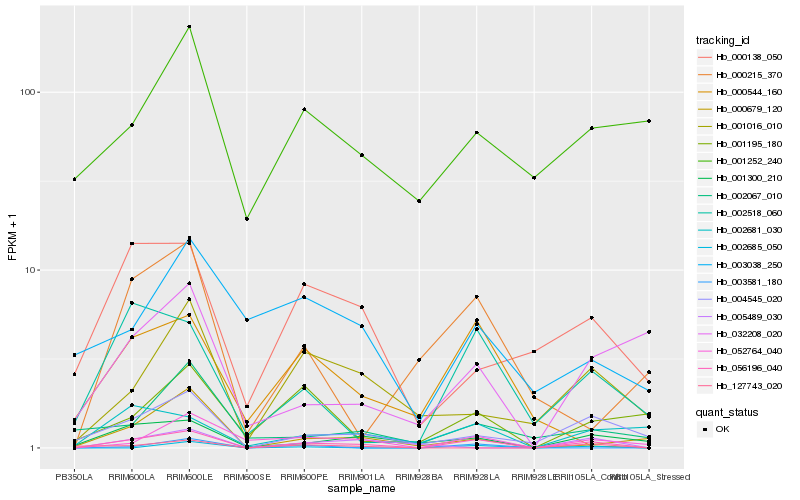
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_056196_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004545_020 |
0.2983273657 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1-like [Populus euphratica] |
| 3 |
Hb_052764_040 |
0.3016917583 |
- |
- |
syntaxin 125 family protein [Populus trichocarpa] |
| 4 |
Hb_001300_210 |
0.3044522151 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 5 |
Hb_002518_060 |
0.3163615421 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |
| 6 |
Hb_000679_120 |
0.3292703431 |
- |
- |
hypothetical protein POPTR_0018s01610g [Populus trichocarpa] |
| 7 |
Hb_001016_010 |
0.3316610146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002681_030 |
0.3402129548 |
- |
- |
PREDICTED: fatty-acid-binding protein 1-like [Jatropha curcas] |
| 9 |
Hb_000138_050 |
0.3472362929 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper protein HAT14, putative [Ricinus communis] |
| 10 |
Hb_000544_160 |
0.3523680052 |
- |
- |
PREDICTED: phosphomethylethanolamine N-methyltransferase-like [Nelumbo nucifera] |
| 11 |
Hb_005489_030 |
0.3605902053 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 12 |
Hb_001195_180 |
0.3612012635 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 13 |
Hb_002067_010 |
0.3621263059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_032208_020 |
0.3639532364 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 15 |
Hb_003581_180 |
0.3657475365 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 16 |
Hb_002685_050 |
0.3687527272 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003038_250 |
0.3768101941 |
- |
- |
PREDICTED: uncharacterized protein LOC105650395 [Jatropha curcas] |
| 18 |
Hb_000215_370 |
0.3773288767 |
- |
- |
myoinositol oxygenase, putative [Ricinus communis] |
| 19 |
Hb_127743_020 |
0.3789773596 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001252_240 |
0.3821980256 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |