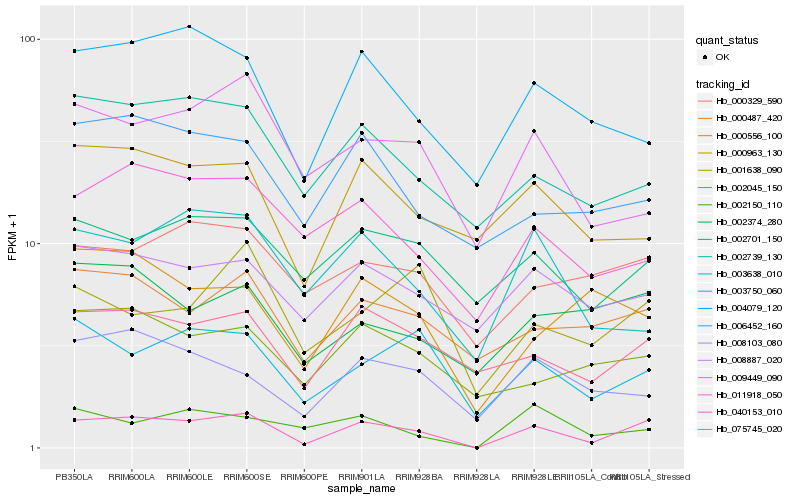
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_040153_010 |
0.0 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 2 |
Hb_075745_020 |
0.1934083313 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003750_060 |
0.2000574783 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000487_420 |
0.2070271773 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |
| 5 |
Hb_004079_120 |
0.2078451204 |
- |
- |
PREDICTED: splicing factor, arginine/serine-rich 19 [Jatropha curcas] |
| 6 |
Hb_002739_130 |
0.2081962665 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 7 |
Hb_011918_050 |
0.208942624 |
- |
- |
- |
| 8 |
Hb_002701_150 |
0.212517142 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_008103_080 |
0.2141200107 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002150_110 |
0.2143473574 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 11 |
Hb_000963_130 |
0.2145318186 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_002045_150 |
0.2157197608 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 13 |
Hb_000329_590 |
0.2158199109 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_008887_020 |
0.215885105 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 15 |
Hb_009449_090 |
0.2160945796 |
- |
- |
PREDICTED: RNA-binding protein 25 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002374_280 |
0.2170330937 |
- |
- |
- |
| 17 |
Hb_006452_160 |
0.2171101544 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 18 |
Hb_001638_090 |
0.2187752784 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 19 |
Hb_003638_010 |
0.2191302732 |
transcription factor |
TF Family: ARID |
PREDICTED: lysine-specific demethylase 5B isoform X2 [Jatropha curcas] |
| 20 |
Hb_000556_100 |
0.2209892311 |
- |
- |
PREDICTED: uncharacterized protein LOC104784782 isoform X2 [Camelina sativa] |