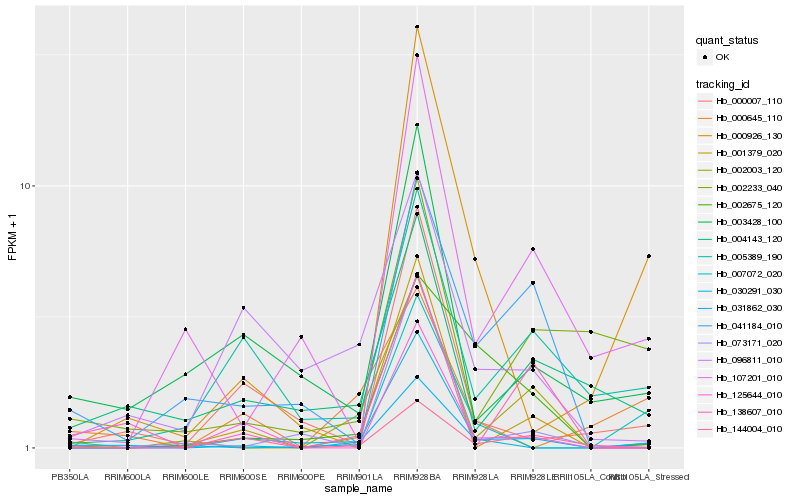
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031862_030 |
0.0 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 2 |
Hb_002003_120 |
0.2550983479 |
- |
- |
PREDICTED: ubiquitin-like-conjugating enzyme ATG10 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001379_020 |
0.2715823532 |
- |
- |
- |
| 4 |
Hb_030291_030 |
0.280870895 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 5 |
Hb_000926_130 |
0.2858748021 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_002675_120 |
0.2859155906 |
- |
- |
DNA-repair protein xrcc1, putative [Ricinus communis] |
| 7 |
Hb_138607_010 |
0.2891367898 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 8 |
Hb_003428_100 |
0.2945380863 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 9 |
Hb_004143_120 |
0.2962567113 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 10 |
Hb_005389_190 |
0.2983155168 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 11 |
Hb_000007_110 |
0.2990471731 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 12 |
Hb_144004_010 |
0.3017989048 |
- |
- |
hypothetical protein JCGZ_01009 [Jatropha curcas] |
| 13 |
Hb_125644_010 |
0.3024441019 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 14 |
Hb_007072_020 |
0.3069031489 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 15 |
Hb_107201_010 |
0.3084595809 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 16 |
Hb_041184_010 |
0.3100784789 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 17 |
Hb_002233_040 |
0.3103311823 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 18 |
Hb_073171_020 |
0.3129635234 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 19 |
Hb_096811_010 |
0.3140344369 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At1g61190 [Vitis vinifera] |
| 20 |
Hb_000645_110 |
0.3145561356 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein RGL1-like [Jatropha curcas] |