| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
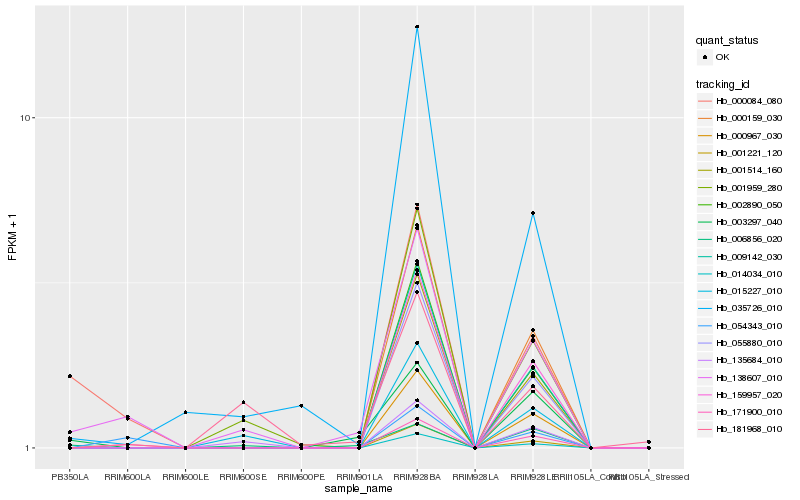
Hb_138607_010 |
0.0 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 2 |
Hb_171900_010 |
0.1949290728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000084_080 |
0.210319082 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001959_280 |
0.2217478626 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 5 |
Hb_015227_010 |
0.2241606636 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_054343_010 |
0.2294539776 |
- |
- |
PREDICTED: uncharacterized protein LOC105793279 [Gossypium raimondii] |
| 7 |
Hb_035726_010 |
0.2303996146 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 8 |
Hb_135684_010 |
0.23623256 |
- |
- |
hypothetical protein [Pseudomonas tolaasii] |
| 9 |
Hb_003297_040 |
0.2385159677 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 10 |
Hb_181968_010 |
0.2434275525 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 11 |
Hb_055880_010 |
0.243704775 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_009142_030 |
0.2438071247 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001514_160 |
0.2438918519 |
- |
- |
hypothetical protein JCGZ_16070 [Jatropha curcas] |
| 14 |
Hb_159957_020 |
0.2441009663 |
- |
- |
- |
| 15 |
Hb_014034_010 |
0.2441567007 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_000159_030 |
0.2442124467 |
- |
- |
putative glycosyltransferase, partial [Aralia elata] |
| 17 |
Hb_002890_050 |
0.2442219978 |
- |
- |
- |
| 18 |
Hb_001221_120 |
0.2444037445 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_000967_030 |
0.2460146587 |
- |
- |
- |
| 20 |
Hb_006856_020 |
0.2491643571 |
- |
- |
- |