| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
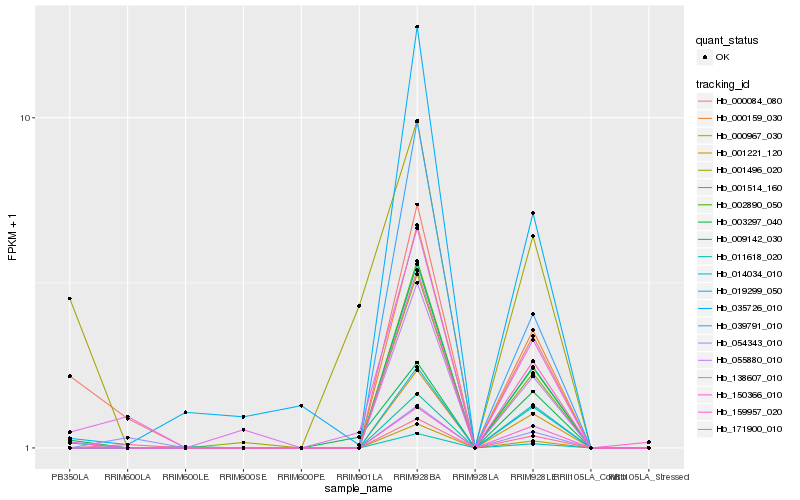
Hb_000084_080 |
0.0 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein, chloroplastic [Jatropha curcas] |
| 2 |
Hb_138607_010 |
0.210319082 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 3 |
Hb_171900_010 |
0.2442010313 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011618_020 |
0.2582317614 |
- |
- |
hypothetical protein OsI_13615 [Oryza sativa Indica Group] |
| 5 |
Hb_150366_010 |
0.2587549175 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 6 |
Hb_001221_120 |
0.2662255805 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_002890_050 |
0.2662990776 |
- |
- |
- |
| 8 |
Hb_014034_010 |
0.2663338631 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_001514_160 |
0.2665484823 |
- |
- |
hypothetical protein JCGZ_16070 [Jatropha curcas] |
| 10 |
Hb_009142_030 |
0.2666596611 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_055880_010 |
0.2668573345 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_159957_020 |
0.2694550848 |
- |
- |
- |
| 13 |
Hb_000159_030 |
0.2697170596 |
- |
- |
putative glycosyltransferase, partial [Aralia elata] |
| 14 |
Hb_000967_030 |
0.2729865954 |
- |
- |
- |
| 15 |
Hb_039791_010 |
0.2734650076 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_003297_040 |
0.2751187694 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 17 |
Hb_054343_010 |
0.2764825928 |
- |
- |
PREDICTED: uncharacterized protein LOC105793279 [Gossypium raimondii] |
| 18 |
Hb_035726_010 |
0.2786513635 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 19 |
Hb_001496_020 |
0.2800972106 |
- |
- |
putative WRKY transcription factor 19 -like protein [Gossypium arboreum] |
| 20 |
Hb_019299_050 |
0.2839364708 |
- |
- |
- |