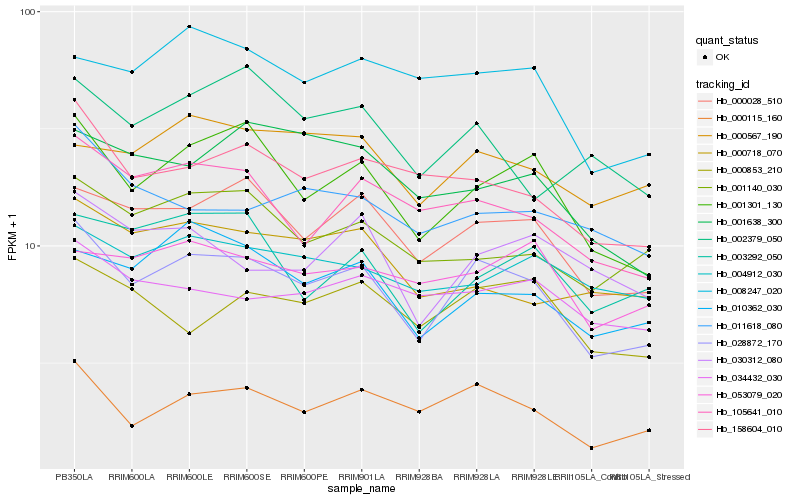
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028872_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001301_130 |
0.081308048 |
- |
- |
PREDICTED: uncharacterized protein LOC105632935 [Jatropha curcas] |
| 3 |
Hb_000028_510 |
0.0845616722 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 4 |
Hb_000115_160 |
0.0849035373 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 5 |
Hb_158604_010 |
0.0990932226 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 6 |
Hb_105641_010 |
0.0998130692 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 7 |
Hb_010362_030 |
0.1048129603 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 8 |
Hb_001140_030 |
0.1132710814 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 9 |
Hb_000853_210 |
0.1150211106 |
- |
- |
PREDICTED: uncharacterized protein LOC105642575 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008247_020 |
0.115409464 |
- |
- |
PREDICTED: cyclin-L1-1-like [Jatropha curcas] |
| 11 |
Hb_030312_080 |
0.118280956 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 12 |
Hb_004912_030 |
0.1183716598 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 13 |
Hb_003292_050 |
0.118664886 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 14 |
Hb_034432_030 |
0.1191487898 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000567_190 |
0.119475595 |
- |
- |
PREDICTED: protein SCAR3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001638_300 |
0.1204459584 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 4 [Jatropha curcas] |
| 17 |
Hb_011618_080 |
0.1217521495 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 18 |
Hb_002379_050 |
0.1225603148 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 19 |
Hb_053079_020 |
0.1227373302 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 20 |
Hb_000718_070 |
0.123226426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |