| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
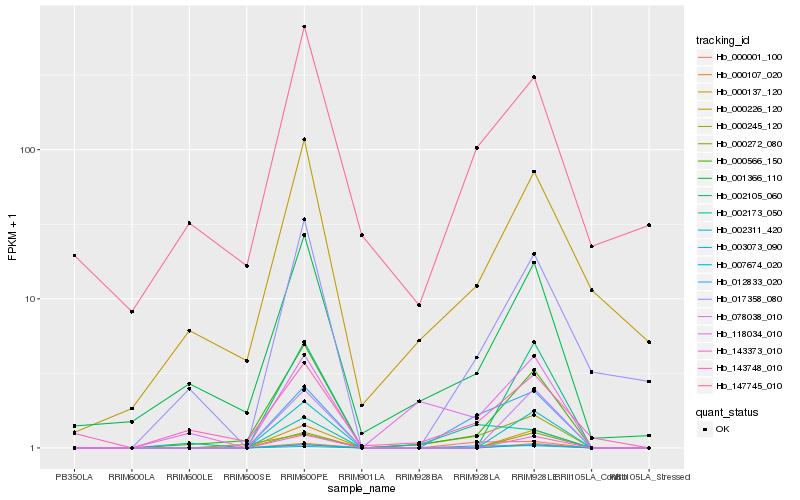
Hb_012833_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000001_100 |
0.2317020543 |
- |
- |
- |
| 3 |
Hb_000566_150 |
0.2595169558 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002173_050 |
0.2648225193 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_017358_080 |
0.2841109042 |
- |
- |
- |
| 6 |
Hb_000245_120 |
0.2933560732 |
- |
- |
hypothetical protein POPTR_0001s18890g [Populus trichocarpa] |
| 7 |
Hb_001366_110 |
0.2946752703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_147745_010 |
0.3040628786 |
- |
- |
PREDICTED: uncharacterized protein LOC105639149 [Jatropha curcas] |
| 9 |
Hb_143748_010 |
0.3053590187 |
- |
- |
PREDICTED: uncharacterized protein LOC105648045 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003073_090 |
0.3063673539 |
transcription factor |
TF Family: B3 |
hypothetical protein POPTR_0004s23920g [Populus trichocarpa] |
| 11 |
Hb_143373_010 |
0.3106614274 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 [Gossypium raimondii] |
| 12 |
Hb_000107_020 |
0.3108448689 |
- |
- |
PREDICTED: uncharacterized protein LOC105032542 [Elaeis guineensis] |
| 13 |
Hb_007674_020 |
0.3112257522 |
- |
- |
PREDICTED: serpin-ZX-like [Jatropha curcas] |
| 14 |
Hb_000272_080 |
0.3114050653 |
- |
- |
temperature-induced lipocalin [Populus tremula x Populus tremuloides] |
| 15 |
Hb_002105_060 |
0.3115005842 |
- |
- |
hypothetical protein POPTR_0022s00320g [Populus trichocarpa] |
| 16 |
Hb_000226_120 |
0.3117534522 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000137_120 |
0.3121679085 |
- |
- |
hypothetical protein JCGZ_15021 [Jatropha curcas] |
| 18 |
Hb_078038_010 |
0.3121937051 |
- |
- |
hypothetical protein CISIN_1g038197mg [Citrus sinensis] |
| 19 |
Hb_002311_420 |
0.312909224 |
- |
- |
hypothetical protein JCGZ_18919 [Jatropha curcas] |
| 20 |
Hb_118034_010 |
0.3130487938 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump [Jatropha curcas] |