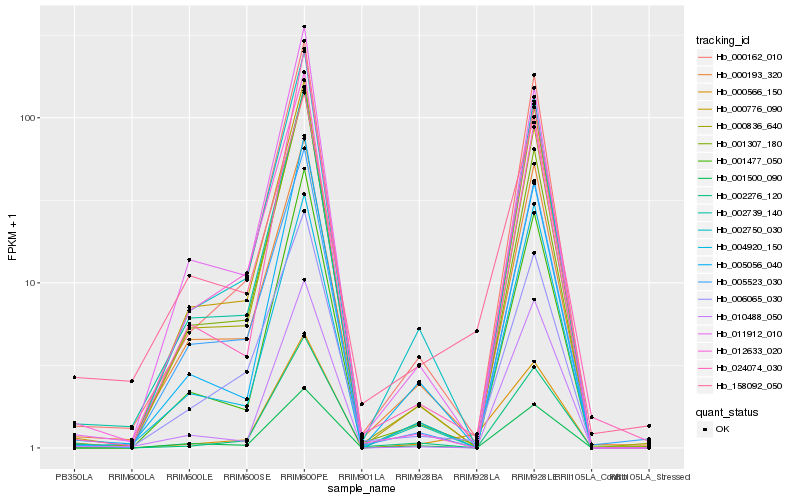
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000566_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000162_010 |
0.1271348282 |
- |
- |
PREDICTED: dirigent protein 23 [Jatropha curcas] |
| 3 |
Hb_002750_030 |
0.1327387787 |
- |
- |
PREDICTED: protein IRX15-LIKE-like [Populus euphratica] |
| 4 |
Hb_006065_030 |
0.1345895123 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000836_640 |
0.1358263548 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 6 |
Hb_001477_050 |
0.1365474607 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_005056_040 |
0.141727292 |
- |
- |
hypothetical protein POPTR_0007s14730g [Populus trichocarpa] |
| 8 |
Hb_000776_090 |
0.1423902203 |
- |
- |
PREDICTED: COBRA-like protein 4 [Jatropha curcas] |
| 9 |
Hb_158092_050 |
0.1431647982 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10 [Jatropha curcas] |
| 10 |
Hb_001500_090 |
0.1433209244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000193_320 |
0.1469350698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_024074_030 |
0.1482514089 |
- |
- |
hypothetical protein CISIN_1g043166mg [Citrus sinensis] |
| 13 |
Hb_002276_120 |
0.1490126369 |
- |
- |
hypothetical protein POPTR_0010s14710g [Populus trichocarpa] |
| 14 |
Hb_001307_180 |
0.1517034213 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |
| 15 |
Hb_011912_010 |
0.1550448614 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_010488_050 |
0.1559089925 |
transcription factor |
TF Family: LIM |
hypothetical protein JCGZ_14428 [Jatropha curcas] |
| 17 |
Hb_004920_150 |
0.1560031193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005523_030 |
0.1560125714 |
- |
- |
PREDICTED: probable galacturonosyltransferase 12 [Jatropha curcas] |
| 19 |
Hb_012633_020 |
0.1564335699 |
- |
- |
laccase, putative [Ricinus communis] |
| 20 |
Hb_002739_140 |
0.1574747581 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |