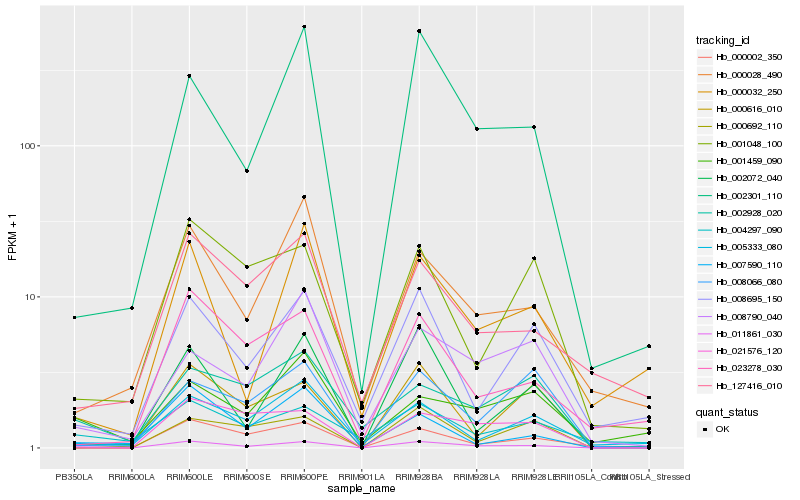
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011861_030 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance protein [Morus notabilis] |
| 2 |
Hb_002301_110 |
0.1989700609 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 3 |
Hb_002072_040 |
0.21915338 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 4 |
Hb_008695_150 |
0.221673306 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000028_490 |
0.2280019413 |
- |
- |
calmodulin-like protein 6a [Populus trichocarpa] |
| 6 |
Hb_008790_040 |
0.2294541317 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG8 [Jatropha curcas] |
| 7 |
Hb_005333_080 |
0.2300445823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_021576_120 |
0.2301062638 |
- |
- |
DNA-directed RNA polymerase subunit, putative [Ricinus communis] |
| 9 |
Hb_002928_020 |
0.231115563 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 10 |
Hb_000032_250 |
0.2331267217 |
- |
- |
PREDICTED: endoglucanase 6 [Jatropha curcas] |
| 11 |
Hb_023278_030 |
0.2332530806 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_127416_010 |
0.23448343 |
- |
- |
LysM domain GPI-anchored protein 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_000616_010 |
0.2347287117 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004297_090 |
0.2352863568 |
- |
- |
PREDICTED: DNA polymerase epsilon catalytic subunit A [Jatropha curcas] |
| 15 |
Hb_008066_080 |
0.2353580686 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 16 |
Hb_000002_350 |
0.2373779333 |
transcription factor |
TF Family: SNF2 |
DNA repair helicase rad5,16, putative [Ricinus communis] |
| 17 |
Hb_001048_100 |
0.237474956 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase [Jatropha curcas] |
| 18 |
Hb_000692_110 |
0.2379185491 |
- |
- |
PREDICTED: NEP1-interacting protein-like 1 [Jatropha curcas] |
| 19 |
Hb_001459_090 |
0.2383641388 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_007590_110 |
0.2386063585 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |