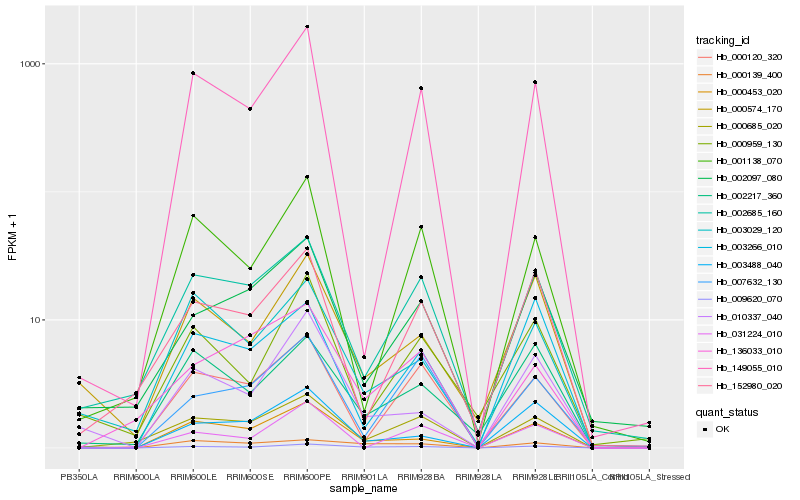
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009620_070 |
0.0 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 2 |
Hb_000139_400 |
0.1646797184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000685_020 |
0.1670709062 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 4 |
Hb_000453_020 |
0.1679744671 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_002217_360 |
0.1838040881 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002685_160 |
0.1918774921 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 7 |
Hb_003488_040 |
0.1936770288 |
transcription factor |
TF Family: NF-YB |
nuclear transcription factor YB7 [Populus nigra x Populus x canadensis] |
| 8 |
Hb_000120_320 |
0.1954239997 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_000574_170 |
0.1960000507 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump-like [Jatropha curcas] |
| 10 |
Hb_010337_040 |
0.2048016393 |
- |
- |
- |
| 11 |
Hb_149055_010 |
0.2061050351 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 12 |
Hb_136033_010 |
0.2100194967 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Gossypium raimondii] |
| 13 |
Hb_007632_130 |
0.2110665613 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003029_120 |
0.2120951967 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 15 |
Hb_002097_080 |
0.2122581423 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 16 |
Hb_001138_070 |
0.2130473122 |
- |
- |
PREDICTED: D-3-phosphoglycerate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_003266_010 |
0.2133287316 |
- |
- |
hypothetical protein JCGZ_21945 [Jatropha curcas] |
| 18 |
Hb_152980_020 |
0.2135367203 |
- |
- |
PREDICTED: protein trichome birefringence-like 33 [Jatropha curcas] |
| 19 |
Hb_000959_130 |
0.2138659644 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 20 |
Hb_031224_010 |
0.2168410733 |
- |
- |
- |