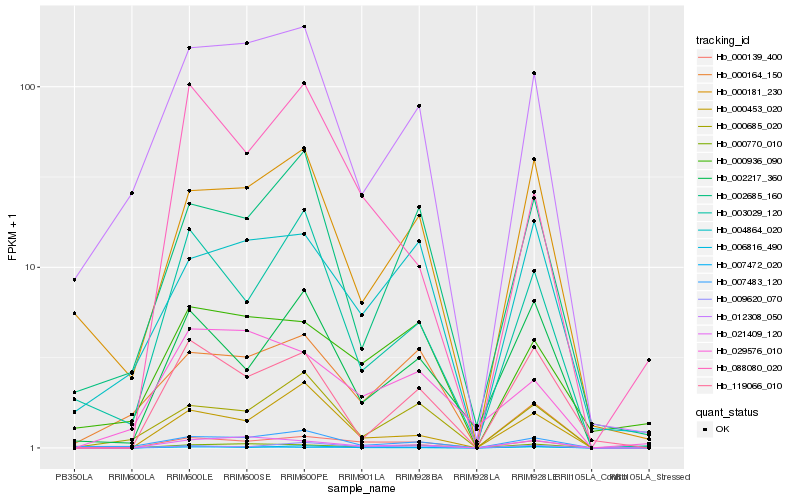
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_400 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009620_070 |
0.1646797184 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 3 |
Hb_000770_010 |
0.1769346641 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 4 |
Hb_007472_020 |
0.202757578 |
- |
- |
hypothetical protein PRUPE_ppa016115mg [Prunus persica] |
| 5 |
Hb_000936_090 |
0.2040905938 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X2 [Jatropha curcas] |
| 6 |
Hb_000453_020 |
0.2059795988 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_021409_120 |
0.2069292816 |
transcription factor |
TF Family: MYB-related |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 8 |
Hb_007483_120 |
0.2086175653 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 9 |
Hb_002217_360 |
0.2140371765 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000685_020 |
0.2146140767 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 11 |
Hb_004864_020 |
0.2158000315 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 12 |
Hb_088080_020 |
0.220630253 |
- |
- |
- |
| 13 |
Hb_003029_120 |
0.2215326649 |
- |
- |
PREDICTED: uncharacterized protein LOC105642638 [Jatropha curcas] |
| 14 |
Hb_012308_050 |
0.2222077782 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 15 |
Hb_029576_010 |
0.2245133091 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31-like [Glycine max] |
| 16 |
Hb_000181_230 |
0.2262263258 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_002685_160 |
0.2294434788 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 18 |
Hb_006816_490 |
0.2324353429 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 21 [Jatropha curcas] |
| 19 |
Hb_119066_010 |
0.2331918683 |
- |
- |
PREDICTED: uncharacterized protein LOC105641552 [Jatropha curcas] |
| 20 |
Hb_000164_150 |
0.2334038726 |
- |
- |
serine/threonine-specific protein kinase, putative [Ricinus communis] |