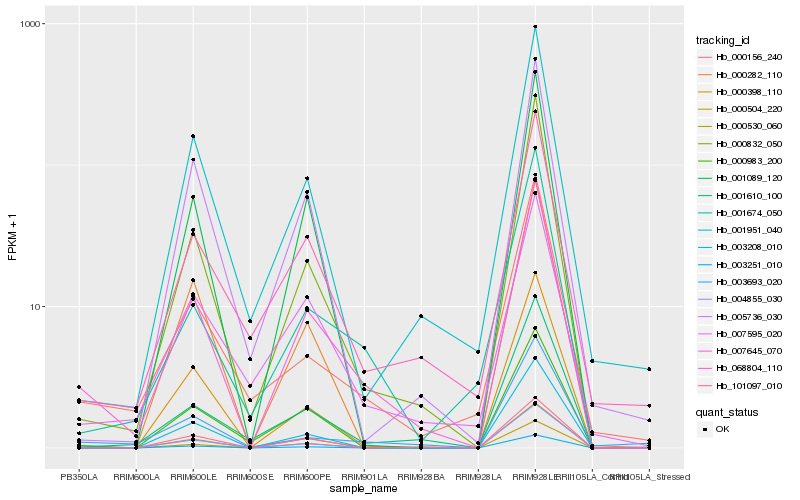
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007645_070 |
0.0 |
- |
- |
photosystem II cytochrome b559 alpha subunit [Cuscuta obtusiflora] |
| 2 |
Hb_000832_050 |
0.1124835146 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Marila laxiflora] |
| 3 |
Hb_001610_100 |
0.1342079893 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_068804_110 |
0.1399591425 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_101097_010 |
0.1429008011 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 6 |
Hb_000983_200 |
0.1431063094 |
- |
- |
PREDICTED: uncharacterized protein LOC105633924 [Jatropha curcas] |
| 7 |
Hb_001089_120 |
0.1432040927 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 8 |
Hb_003251_010 |
0.1462222472 |
- |
- |
hypothetical protein MIMGU_mgv1a025126mg [Erythranthe guttata] |
| 9 |
Hb_001951_040 |
0.1464929442 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000156_240 |
0.1467151233 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 11 |
Hb_000398_110 |
0.1480520104 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 4-like [Populus euphratica] |
| 12 |
Hb_003208_010 |
0.1483987427 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 13 |
Hb_001674_050 |
0.1484934275 |
- |
- |
cytochrome b6 (chloroplast) [Apios americana] |
| 14 |
Hb_007595_020 |
0.1485022064 |
- |
- |
maturase K [Hevea brasiliensis] |
| 15 |
Hb_004855_030 |
0.1493207722 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 16 |
Hb_000504_220 |
0.1495017192 |
- |
- |
hypothetical protein JCGZ_10249 [Jatropha curcas] |
| 17 |
Hb_000530_060 |
0.1501633467 |
- |
- |
- |
| 18 |
Hb_005736_030 |
0.1503206519 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 19 |
Hb_003693_020 |
0.1503832053 |
- |
- |
Ycf1, partial (chloroplast) [Clusia rosea] |
| 20 |
Hb_000282_110 |
0.1516502387 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein POPTR_0006s24560g [Populus trichocarpa] |