| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
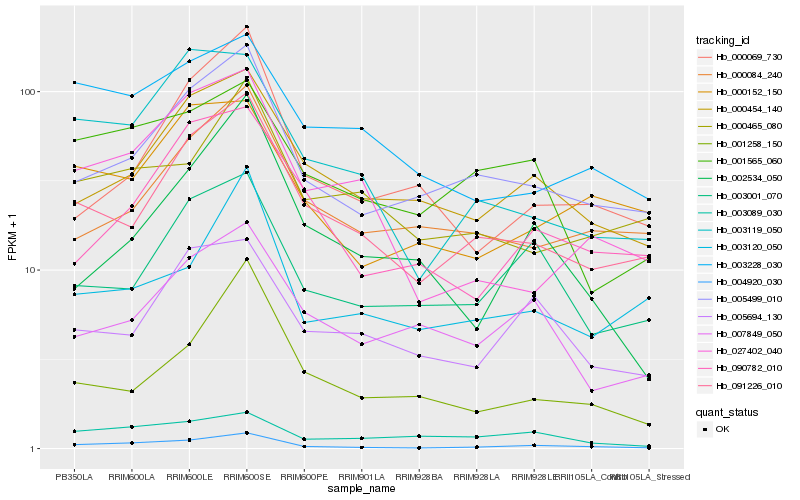
Hb_004920_030 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase CKI1 [Jatropha curcas] |
| 2 |
Hb_005499_010 |
0.152346405 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 3 |
Hb_091226_010 |
0.1599485794 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 4 |
Hb_003119_050 |
0.1695050455 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_027402_040 |
0.171653083 |
- |
- |
Thioredoxin II, putative [Ricinus communis] |
| 6 |
Hb_000454_140 |
0.176820983 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 7 |
Hb_000152_150 |
0.1802879212 |
- |
- |
PREDICTED: uncharacterized protein LOC105648558 [Jatropha curcas] |
| 8 |
Hb_003001_070 |
0.1821779365 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |
| 9 |
Hb_090782_010 |
0.1822262573 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 10 |
Hb_000069_730 |
0.1865578982 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001565_060 |
0.1890658707 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_003228_030 |
0.1918574256 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_001258_150 |
0.1925517796 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 14 |
Hb_005694_130 |
0.192673095 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 15 |
Hb_002534_050 |
0.1942577433 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 16 |
Hb_003120_050 |
0.1946210332 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 17 |
Hb_003089_030 |
0.1953585761 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000084_240 |
0.1957228876 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 19 |
Hb_000465_080 |
0.1977539214 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 20 |
Hb_007849_050 |
0.1983255031 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |