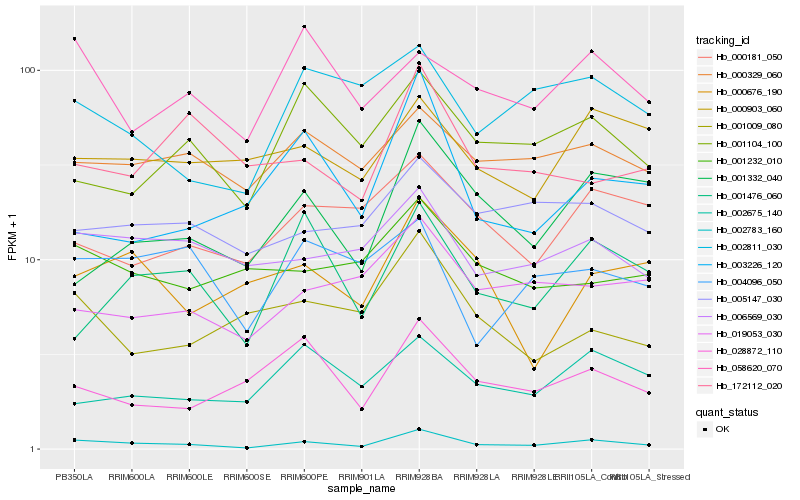
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_160 |
0.0 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 2 |
Hb_006569_030 |
0.1913761744 |
- |
- |
PREDICTED: uncharacterized protein LOC105650907 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000329_060 |
0.2010148429 |
- |
- |
chloroplast 5-enolpyruvylshikimate 3-phosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_001476_060 |
0.2015163007 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 5 |
Hb_028872_110 |
0.2028446028 |
- |
- |
PREDICTED: uncharacterized protein LOC105644776 [Jatropha curcas] |
| 6 |
Hb_001332_040 |
0.2037963564 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |
| 7 |
Hb_005147_030 |
0.2039952204 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 8 |
Hb_001009_080 |
0.2042423059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002811_030 |
0.205293309 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 10 |
Hb_019053_030 |
0.2055507731 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 11 |
Hb_000676_190 |
0.2073475769 |
- |
- |
hypothetical protein JCGZ_08279 [Jatropha curcas] |
| 12 |
Hb_001104_100 |
0.2073672699 |
- |
- |
RecName: Full=Enolase 2; AltName: Full=2-phospho-D-glycerate hydro-lyase 2; AltName: Full=2-phosphoglycerate dehydratase 2; AltName: Allergen=Hev b 9 [Hevea brasiliensis] |
| 13 |
Hb_000903_060 |
0.2076702513 |
- |
- |
PREDICTED: ras-related protein RABA5e [Jatropha curcas] |
| 14 |
Hb_000181_050 |
0.2080170554 |
- |
- |
PREDICTED: uncharacterized protein LOC105637463 [Jatropha curcas] |
| 15 |
Hb_004096_050 |
0.2085712801 |
- |
- |
hypothetical protein B456_006G229700 [Gossypium raimondii] |
| 16 |
Hb_003226_120 |
0.2100499657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001232_010 |
0.2105426785 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 18 |
Hb_002675_140 |
0.2114337468 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 19 |
Hb_058620_070 |
0.2120895485 |
- |
- |
unnamed protein product [Chondrus crispus] |
| 20 |
Hb_172112_020 |
0.2124907059 |
- |
- |
PREDICTED: fumarylacetoacetase [Jatropha curcas] |