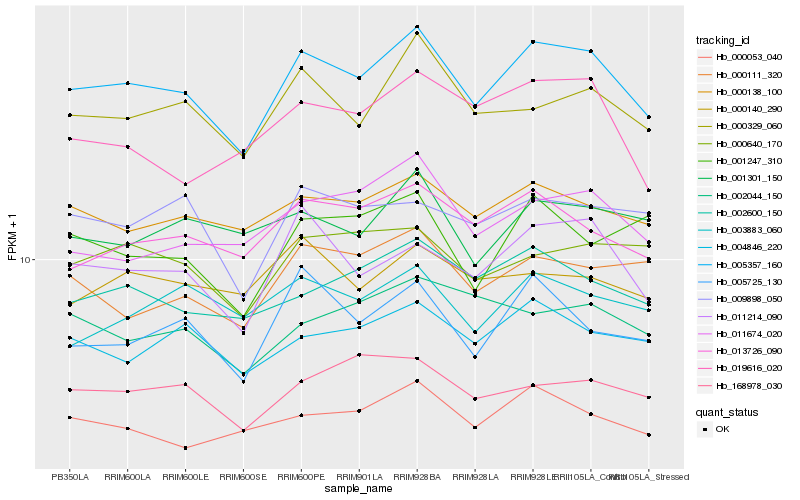
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005357_160 |
0.0 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 2 |
Hb_011214_090 |
0.0655041352 |
- |
- |
PREDICTED: aspartate carbamoyltransferase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000329_060 |
0.0704231733 |
- |
- |
chloroplast 5-enolpyruvylshikimate 3-phosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_000138_100 |
0.072351736 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 5 |
Hb_013726_090 |
0.0728058707 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 6 |
Hb_004846_220 |
0.0750214514 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000053_040 |
0.0750837777 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 8 |
Hb_002600_150 |
0.0757187041 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005725_130 |
0.0770007606 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 10 |
Hb_011674_020 |
0.0793237045 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 regulatory subunit-like [Jatropha curcas] |
| 11 |
Hb_168978_030 |
0.0796136587 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 12 |
Hb_000640_170 |
0.080738173 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000111_320 |
0.0807647277 |
transcription factor |
TF Family: bZIP |
Transcription factor HBP-1b(c1), putative [Ricinus communis] |
| 14 |
Hb_009898_050 |
0.0827342777 |
- |
- |
PREDICTED: beta-taxilin [Jatropha curcas] |
| 15 |
Hb_001301_150 |
0.0837400973 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 16 |
Hb_003883_060 |
0.0847403181 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_019616_020 |
0.0847812208 |
- |
- |
PREDICTED: alanine--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_000140_290 |
0.0856501988 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 59 kDa protein isoform X1 [Populus euphratica] |
| 19 |
Hb_002044_150 |
0.0874861263 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 20 |
Hb_001247_310 |
0.0877409788 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s00450g [Populus trichocarpa] |