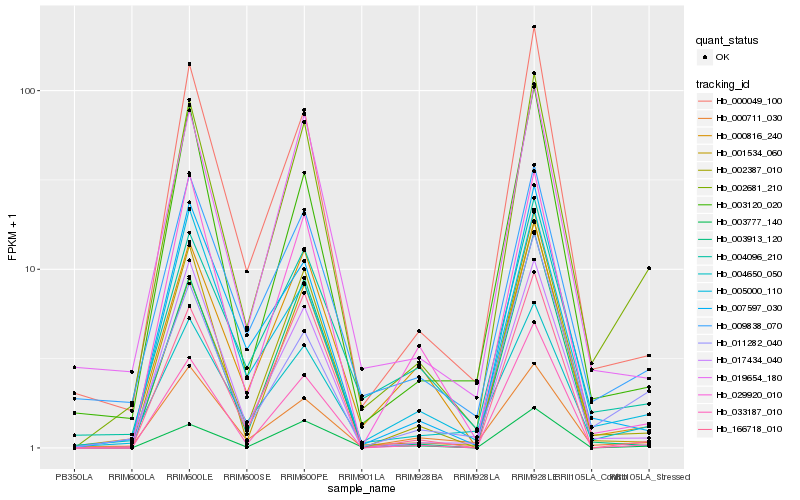
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002681_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633686 [Jatropha curcas] |
| 2 |
Hb_004650_050 |
0.1138627578 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_019654_180 |
0.1204326938 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 4 |
Hb_000049_100 |
0.1216455185 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 5 |
Hb_017434_040 |
0.1259389298 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670-like [Jatropha curcas] |
| 6 |
Hb_009838_070 |
0.1261315738 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003777_140 |
0.1261378006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_033187_010 |
0.1357614659 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 9 |
Hb_011282_040 |
0.1360777906 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 10 |
Hb_001534_060 |
0.1362157738 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 11 |
Hb_003120_020 |
0.138921698 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 13-like [Prunus mume] |
| 12 |
Hb_005000_110 |
0.1390045016 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000711_030 |
0.1399231701 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 14 |
Hb_004096_210 |
0.1402507601 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 15 |
Hb_029920_010 |
0.1420423258 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 16 |
Hb_007597_030 |
0.1423866278 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_002387_010 |
0.1427934615 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000816_240 |
0.1436485666 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF2, chloroplastic [Jatropha curcas] |
| 19 |
Hb_166718_010 |
0.1436648321 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 20 |
Hb_003913_120 |
0.1443427369 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |