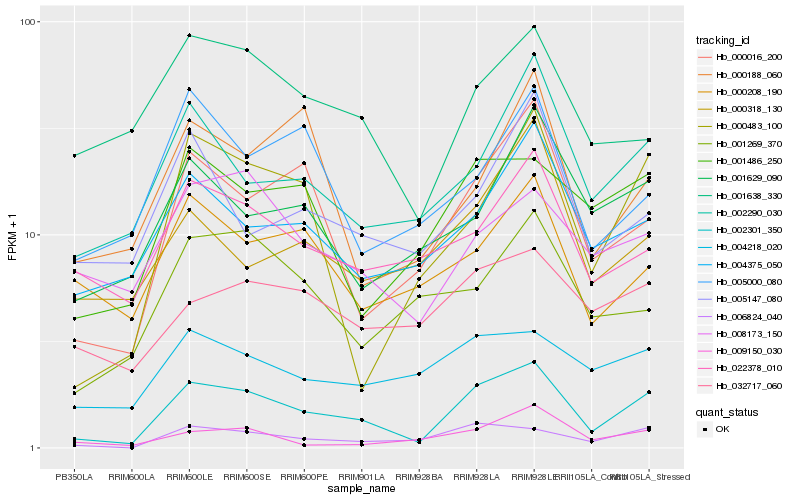
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002301_350 |
0.0 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 2 |
Hb_006824_040 |
0.1731582283 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Populus euphratica] |
| 3 |
Hb_000016_200 |
0.181352308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000483_100 |
0.1929884659 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 9-like [Jatropha curcas] |
| 5 |
Hb_002290_030 |
0.1985307799 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001486_250 |
0.2046295329 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 7 |
Hb_004375_050 |
0.204848824 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_001638_330 |
0.2121135048 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_022378_010 |
0.2126717209 |
- |
- |
PREDICTED: uncharacterized protein LOC105634715 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004218_020 |
0.2145829801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008173_150 |
0.21572271 |
- |
- |
PREDICTED: uncharacterized protein LOC105645812 [Jatropha curcas] |
| 12 |
Hb_001629_090 |
0.2171771138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_009150_030 |
0.2177060319 |
- |
- |
hypothetical protein POPTR_0010s14920g [Populus trichocarpa] |
| 14 |
Hb_005147_080 |
0.2195720621 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_032717_060 |
0.221232021 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000208_190 |
0.2217721834 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 17 |
Hb_001269_370 |
0.222603956 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 18 |
Hb_000188_060 |
0.2229910006 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 4-like [Jatropha curcas] |
| 19 |
Hb_005000_080 |
0.2243114732 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 20 |
Hb_000318_130 |
0.2247851477 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |